7XSF
 
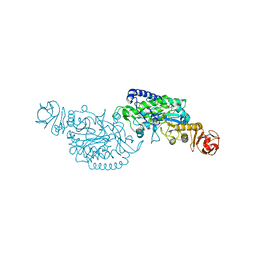 | | Crystal structure of ClAgl29A | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSG
 
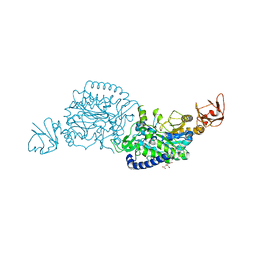 | | Crystal structure of ClAgl29B | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
7XSH
 
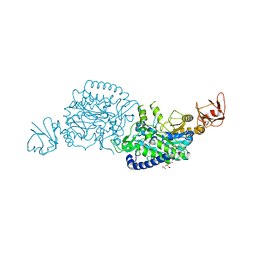 | | Crystal structure of ClAgl29B bound with L-glucose | | Descriptor: | Alpha-L-fucosidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shishiuchi, R, Kang, H, Tagami, T, Okuyama, M. | | Deposit date: | 2022-05-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Discovery of alpha-l-Glucosidase Raises the Possibility of alpha-l-Glucosides in Nature.
Acs Omega, 7, 2022
|
|
2ZK9
 
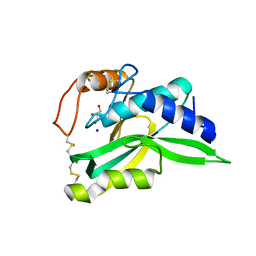 | | Crystal Structure of Protein-glutaminase | | Descriptor: | GLYCEROL, Protein-glutaminase, SODIUM ION | | Authors: | Hashizume, R. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-17 | | Last modified: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of protein glutaminase and its pro forms converted into enzyme-substrate complex
J.Biol.Chem., 286, 2011
|
|
3A56
 
 | |
2YVU
 
 | |
2YVT
 
 | |
1XQD
 
 | | Crystal structure of P450NOR complexed with 3-pyridinealdehyde adenine dinucleotide | | Descriptor: | CYTOCHROME P450 55A1, NICOTINIC ACID ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Takaya, N, Su, F, Wakagi, T, Shoun, H. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1ULW
 
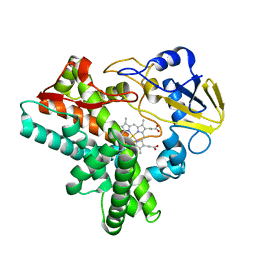 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1WW1
 
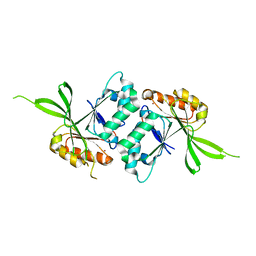 | |
2E7Y
 
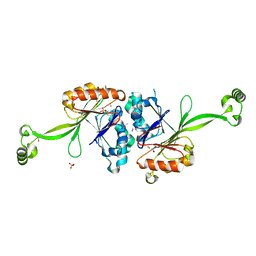 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2EY4
 
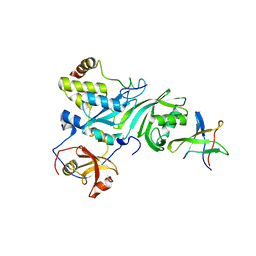 | | Crystal Structure of a Cbf5-Nop10-Gar1 Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, ZINC ION, ... | | Authors: | Rashid, R, Liang, B, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita.
Mol.Cell, 21, 2006
|
|
6U0S
 
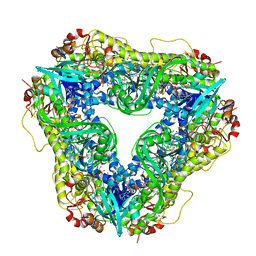 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6U0P
 
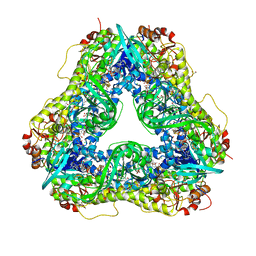 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6UQV
 
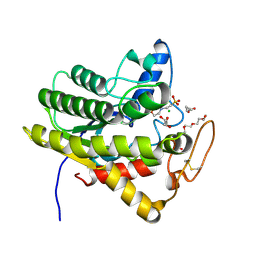 | | Crystal structure of ChoE, a bacterial acetylcholinesterase from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BUTANOIC ACID, CHLORIDE ION, ... | | Authors: | Shi, R, Pham, V.D, To, T.A. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
5CPC
 
 | |
5CQ9
 
 | |
7KEZ
 
 | |
7KF1
 
 | |
7KF0
 
 | |
6M9M
 
 | |
6MXR
 
 | |
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6MXS
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98F,HC-G99M] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Shi, R, Picard, M.-E, Manenda, M.S. | | Deposit date: | 2018-10-31 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6MY5
 
 | |
