8QIC
 
 | |
5K2H
 
 | |
5K2F
 
 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
5WHN
 
 | | Crystal structure of the segment, NFGAFS, from the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WIQ
 
 | | Crystal structure of the segment, GFNGGFG, from the low complexity domain of TDP-43, residues 396-402 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WHP
 
 | | Crystal structure of the segment, NFGTFS, from the A315T familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | Segment of TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WKB
 
 | | MicroED structure of the segment, NFGEFS, from the A315E familial variant of the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-07-24 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ME8
 
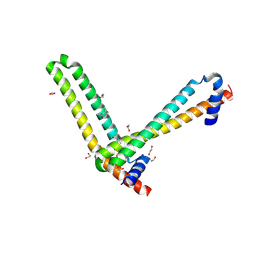 | | N-terminal domain of the human tumor suppressor ING5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Inhibitor of growth protein 5 | | Authors: | Roversi, P, Blanco, F.J, Rojas, A.L, Buitrago, J.A.R. | | Deposit date: | 2016-11-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
5MTO
 
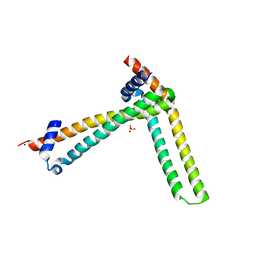 | | N-terminal domain of the human tumor suppressor ING5 C19S mutant | | Descriptor: | Inhibitor of growth protein 5, SODIUM ION, SULFATE ION | | Authors: | Ormaza, G, Buitrago, J.A.R, Roversi, P, Rojas, A.L, Blanco, F.J. | | Deposit date: | 2017-01-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
8DZZ
 
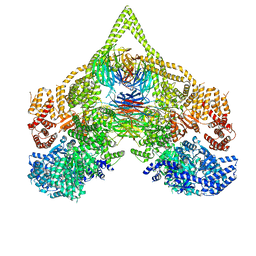 | | Cryo-EM structure of chi dynein bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E00
 
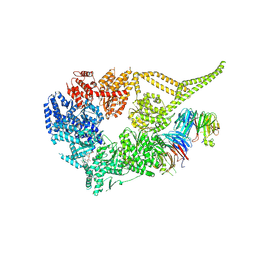 | | Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7N2L
 
 | | MicroED structure of a mutant mammalian prion segment | | Descriptor: | prion protein | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2K
 
 | | MicroED structure of sequence variant of repeat segment of the yeast prion New1p phased by ARCIMBOLDO-BORGES | | Descriptor: | prion New1p | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.301 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2H
 
 | | X-Ray structure of a sequence variant of a repeat segment of the yeast prion New1p | | Descriptor: | prion New1p | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2G
 
 | | MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.201 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2J
 
 | | MicroED structure of a mutant mammalian prion segment phased by ARCIMBOLDO-BORGES | | Descriptor: | prion protein | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2F
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2D
 
 | | MicroED structure of human zinc finger protein 292 segment (534-542) phased by ARCIMBOLDO-BORGES | | Descriptor: | DIMETHYL SULFOXIDE, zinc finger protein 292 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.503 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2E
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2I
 
 | | MicroED structure of human LECT2 (45-53) phased by ARCIMBOLDO-BORGES | | Descriptor: | LECT2 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
6PQ5
 
 | |
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7R1E
 
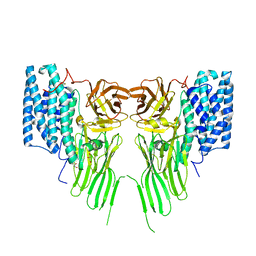 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
