1SOA
 
 | | Human DJ-1 with sulfinic acid | | Descriptor: | RNA-binding protein regulatory subunit; oncogene DJ1 | | Authors: | Canet-Aviles, R, Wilson, M.A, Miller, D.W, Ahmad, R, McLendon, C, Bandyopadhyay, S, Baptista, M.J, Ringe, D, Petsko, G.A, Cookson, M.R. | | Deposit date: | 2004-03-13 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2GMT
 
 | | THREE-DIMENSIONAL STRUCTURE OF CHYMOTRYPSIN INACTIVATED WITH (2S) N-ACETYL-L-ALANYL-L-PHENYLALANYL-CHLOROETHYL KETONE: IMPLICATIONS FOR THE MECHANISM OF INACTIVATION OF SERINE PROTEASES BY CHLOROKETONES | | Descriptor: | (2S) N-ACETYL-L-ALANYL-ALPHAL-PHENYLALANYL-CHLOROETHYLKETONE, GAMMA-CHYMOTRYPSIN | | Authors: | Kreutter, K, Steinmetz, A.C.U, Liang, T.-C, Prorok, M, Abeles, R, Ringe, D. | | Deposit date: | 1994-09-07 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of chymotrypsin inactivated with (2S)-N-acetyl-L-alanyl-L-phenylalanyl alpha-chloroethane: implications for the mechanism of inactivation of serine proteases by chloroketones.
Biochemistry, 33, 1994
|
|
3DHB
 
 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
1XYA
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
4GCH
 
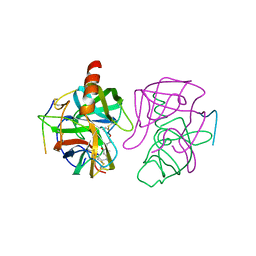 | |
1XYB
 
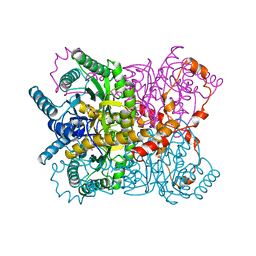 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYC
 
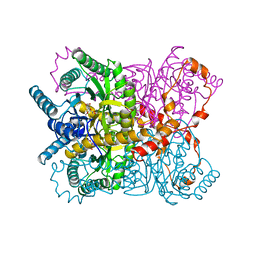 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | Descriptor: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-01-03 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
4N0B
 
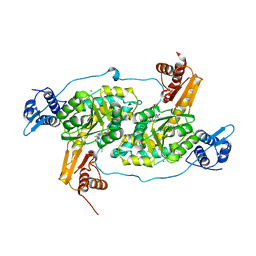 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MGR
 
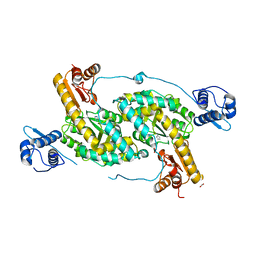 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3DAA
 
 | |
3PAA
 
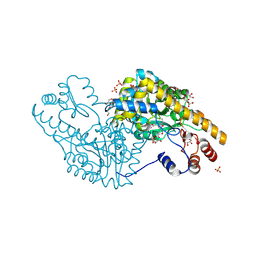 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 8.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
3PA9
 
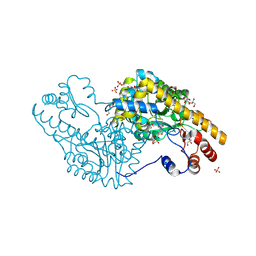 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
1ASF
 
 | |
1ASB
 
 | |
1AAM
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1ASD
 
 | |
1ASC
 
 | |
1G2W
 
 | | E177S MUTANT OF THE PYRIDOXAL-5'-PHOSPHATE ENZYME D-AMINO ACID AMINOTRANSFERASE | | Descriptor: | ACETATE ION, D-ALANINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lepore, B.W, Ringe, D. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on an Active Site Residue, E177, That Affects Binding of the Coenzyme in D-Amino Acid Transaminase, and Mechanistic Studies on a Suicide Substrate
Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins, 10th Annual International Symposium on Vitamin B6, 2000
|
|
1ASG
 
 | |
1ASE
 
 | |
1AAW
 
 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
5T4J
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UGQ
 
 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5UOH
 
 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
