8XV1
 
 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUY
 
 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
2DWM
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D3V
 
 | | Crystal Structure of Leukocyte Ig-like Receptor A5 (LILRA5/LIR9/ILT11) | | Descriptor: | leukocyte immunoglobulin-like receptor subfamily A member 5 isoform 1 | | Authors: | Shiroishi, M, Kajikawa, M, Kuroki, K, Ose, T, Kohda, D, Maenaka, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the human monocyte-activating receptor,
J.Biol.Chem., 281, 2006
|
|
3A3Q
 
 | | Structure of N59D HEN EGG-WHITE LYSOZYME in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
3WMD
 
 | | Crystal structure of epoxide hydrolase MonBI | | Descriptor: | Probable monensin biosynthesis isomerase | | Authors: | Minami, A, Ose, T, Sato, K, Oikawa, A, Kuroki, K, Maenaka, K, Oguri, H, Oikawa, H. | | Deposit date: | 2013-11-16 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Allosteric regulation of epoxide opening cascades by a pair of epoxide hydrolases in monensin biosynthesis
Acs Chem.Biol., 9, 2014
|
|
3A3R
 
 | | Structure of N59D HEN EGG-WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
3WV0
 
 | | O-glycan attached to herpes simplex virus type 1 glycoprotein gB is recognized by the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | Envelope glycoprotein B, N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-galactopyranose, Paired immunoglobulin-like type 2 receptor alpha, ... | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WUZ
 
 | | Crystal structure of the Ig V-set domain of human paired immunoglobulin-like type 2 receptor alpha | | Descriptor: | CITRIC ACID, ISOPROPYL ALCOHOL, Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Kuroki, K, Wang, J, Ose, T, Yamaguchi, M, Tabata, S, Maita, N, Nakamura, S, Kajikawa, M, Kogure, A, Satoh, T, Arase, H, Maenaka, K. | | Deposit date: | 2014-05-10 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILR alpha
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WH3
 
 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WH2
 
 | | Human Mincle in complex with citrate | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ALW
 
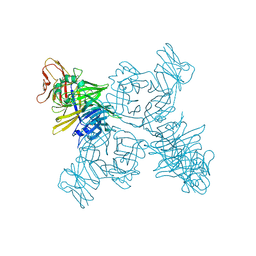 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALX
 
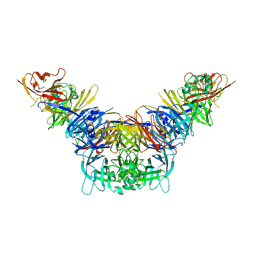 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (MV-H(L482R)-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,LINKER,CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALZ
 
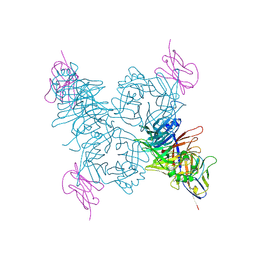 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CDw150, Hemagglutinin | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.515 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3WO9
 
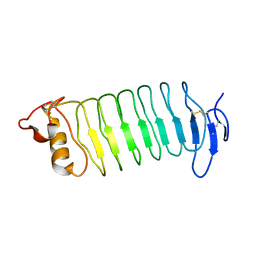 | | Crystal structure of the lamprey variable lymphocyte receptor C | | Descriptor: | Variable lymphocyte receptor C | | Authors: | Kanda, R, Sutoh, Y, Kasamatsu, J, Maenaka, K, Kasahara, M, Ose, T. | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module.
Plos One, 9, 2014
|
|
1LZR
 
 | |
1LZS
 
 | |
8YYU
 
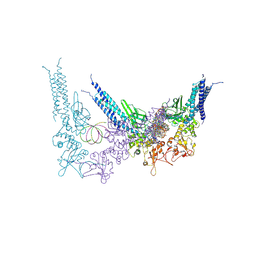 | | A tetrameric STAT1-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*TP*G)-3'), Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Sugiyama, A, Minami, M, Sugita, Y, Ose, T. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein.
Sci.Signal., 18, 2025
|
|
8YYV
 
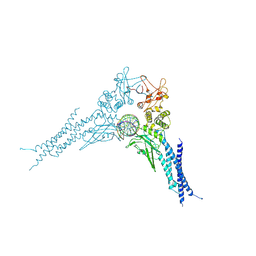 | | A dimeric STAT1-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*TP*G)-3'), Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Sugiyama, A, Minami, M, Sugita, Y, Ose, T. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural analysis reveals how tetrameric tyrosine-phosphorylated STAT1 is targeted by the rabies virus P-protein.
Sci.Signal., 18, 2025
|
|
9L3S
 
 | | Staphylococcus aureus lipase-Penfluridol complex (in space) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamo, M, Furubayasi, N, Inaka, K, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-19 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
9L3C
 
 | | Staphylococcus aureus lipase-Penfluridol complex (on the ground) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
7XBD
 
 | | Cryo-EM structure of human galanin receptor 2 | | Descriptor: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ishimoto, N, Kita, S, Park, S.Y. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
7CEE
 
 | | Crystal structure of mouse neuroligin-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-3 | | Authors: | Yamagata, A, Yoshida, T, Shiroshima, T, Maeda, A, Fukai, S. | | Deposit date: | 2020-06-23 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Canonical versus non-canonical transsynaptic signaling of neuroligin 3 tunes development of sociality in mice.
Nat Commun, 12, 2021
|
|
