5YLO
 
 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
5WQE
 
 | |
5WTJ
 
 | | Crystal structure of an endonuclease | | Descriptor: | CRISPR-associated endoribonuclease C2c2 | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2016-12-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
8WZ4
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 (localized refinement) | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV Fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 (localized refinement)
To Be Published
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7BWD
 
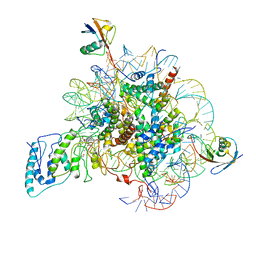 | | Structure of Dot1L-H2BK34ub Nucleosome Complex | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lou, Z.Y, Liu, L, Cao, L, Ai, H.S, Sun, Z.X. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structure of the Dot1L-H2BK34ub Nucleosome Complex Reveals an Unprecedented Crosstalk Activation Mechanism through Nucleosome Shape Distortion
To Be Published
|
|
7T42
 
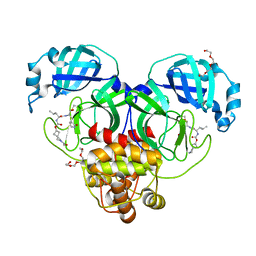 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T43
 
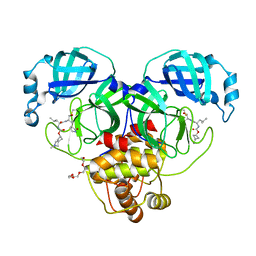 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T44
 
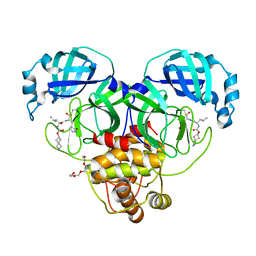 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7SU3
 
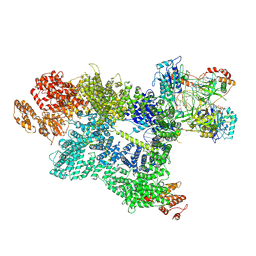 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SUD
 
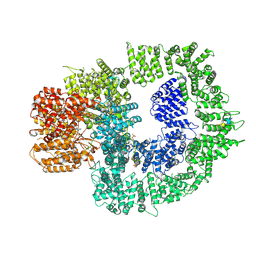 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
2VL7
 
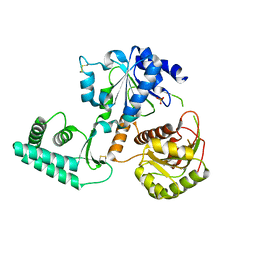 | | Structure of S. tokodaii Xpd4 | | Descriptor: | PHOSPHATE ION, XPD | | Authors: | Naismith, J.H, Johnson, K.A, Oke, M, McMahon, S.A, Liu, L, White, M.F, Zawadski, M, Carter, L.G. | | Deposit date: | 2008-01-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the DNA Repair Helicase Xpd.
Cell(Cambridge,Mass.), 133, 2008
|
|
8CZV
 
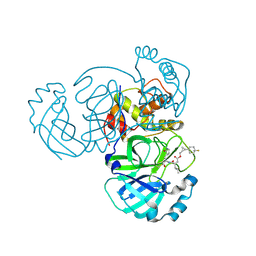 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8E6D
 
 | | Crystal structure of MERS 3CL protease in complex with a p-fluorophenyl dimethyl sulfane inhibitor | | Descriptor: | (1R,2S)-2-{[N-({2-[(4-fluorophenyl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6B
 
 | | Crystal structure of MERS 3CL protease in complex with a dimethyl sulfinyl benzene inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-2-[[2-methyl-2-[oxidanyl(phenyl)-$l^{3}-sulfanyl]propoxy]carbonylamino]pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid, N~2~-(ethoxycarbonyl)-N-{(1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6E
 
 | | Crystal structure of MERS 3CL protease in complex with a phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[2-(phenylsulfanyl)ethoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 2-phenylsulfanylethyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, Orf1a protein | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Madden, T.K, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6C
 
 | | Crystal structure of MERS 3CL protease in complex with a m-fluorophenyl dimethyl sulfane inhibitor | | Descriptor: | Orf1a protein, [2-(3-fluorophenyl)sulfanyl-2-methyl-propyl] ~{N}-[(2~{S})-1-[[3-[(3~{S})-2-$l^{3}-oxidanylidenepyrrolidin-3-yl]-1-$l^{1}-oxidanylsulfonyl-1-oxidanyl-propan-2-yl]-$l^{2}-azanyl]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
6C4X
 
 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4Z
 
 | |
