3EFD
 
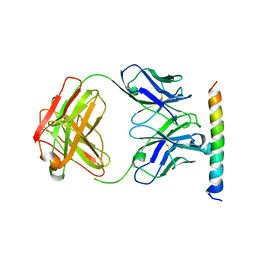 | | The crystal structure of the cytoplasmic domain of KcsA | | Descriptor: | FabH, FabL, KcsA | | Authors: | Uysal, S, Vasquez, V, Tereshko, V, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EFF
 
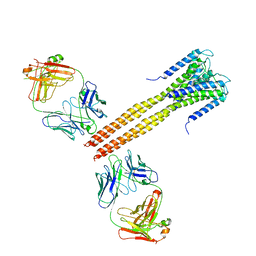 | | The Crystal Structure of Full-Length KcsA in its Closed Conformation | | Descriptor: | FAB, Voltage-gated potassium channel | | Authors: | Uysal, S, Vasquez, V, Tereshko, T, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4JMH
 
 | |
4JMG
 
 | |
3IVK
 
 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
2IQY
 
 | | Rat Phosphatidylethanolamine-Binding Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Heil, G.L, Koide, S, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Rat Phosphatidylethanolamine-Binding Protein
To be Published
|
|
2OY8
 
 | |
2OY7
 
 | |
2QBW
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | PDZ-Fibronectin fusion protein, Polypeptide | | Authors: | Huang, J, Makabe, K, Koide, A, Koide, S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of protein function leaps by directed domain interface evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2OBG
 
 | |
2OYB
 
 | |
2R8S
 
 | | High resolution structure of a specific synthetic FAB bound to P4-P6 RNA ribozyme domain | | Descriptor: | Fab heavy chain, Fab light chain, MAGNESIUM ION, ... | | Authors: | Ye, J.D, Tereshko, V, Sidhu, S.S, Koide, S, Kossiakoff, A.A, Piccirilli, J.A. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic antibodies for specific recognition and crystallization of structured RNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CKA
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A, PENTAETHYLENE GLYCOL | | Authors: | Makabe, K, Biancalana, M, Terechko, V, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Minimalist design of water-soluble cross-{beta} architecture.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3CKF
 
 | | The crystal structure of OspA deletion mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The promiscuity of beta-strand pairing allows for rational design of beta-sheet face inversion
J.Am.Chem.Soc., 130, 2008
|
|
3CKG
 
 | | The crystal structure of OspA deletion mutant | | Descriptor: | MAGNESIUM ION, Outer surface protein A | | Authors: | Makabe, K, Koide, S. | | Deposit date: | 2008-03-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The promiscuity of beta-strand pairing allows for rational design of beta-sheet face inversion
J.Am.Chem.Soc., 130, 2008
|
|
3EEX
 
 | | The crystal structure of OspA mutant | | Descriptor: | HEXAETHYLENE GLYCOL, Outer Surface Protein A | | Authors: | Makabe, K, Biancalana, M, Koide, S. | | Deposit date: | 2008-09-06 | | Release date: | 2009-09-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Minimalist design of water-soluble cross-{beta} architecture.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2OY5
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Biancalana, M, Yan, S, Koide, S. | | Deposit date: | 2007-02-21 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aromatic cluster mutations produce focal modulations of beta-sheet structure.
Protein Sci., 24, 2015
|
|
2OL7
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2OL6
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2OL8
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-01-18 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2OY1
 
 | | The crystal structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Makabe, K, Terechko, V, Koide, S. | | Deposit date: | 2007-02-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | beta-Strand Flipping and Slipping Triggered by Turn Replacement Reveal the Opportunistic Nature of beta-Strand Pairing
J.Am.Chem.Soc., 129, 2007
|
|
2PI3
 
 | |
8SLZ
 
 | |
3K2M
 
 | |
6TLC
 
 | | Unphosphorylated human STAT3 in complex with MS3-6 monobody | | Descriptor: | Monobody, Signal transducer and activator of transcription 3 | | Authors: | La Sala, G, Lau, K, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Selective inhibition of STAT3 signaling using monobodies targeting the coiled-coil and N-terminal domains.
Nat Commun, 11, 2020
|
|
