8D1O
 
 | | hBest1_345 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1F
 
 | | hBest2 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1N
 
 | | bBest2_345 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1J
 
 | | hBest1 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1E
 
 | | hBest2 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.78 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1L
 
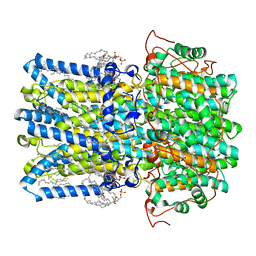 | | hBest1 Ca2+-bound partially open aperture state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1H
 
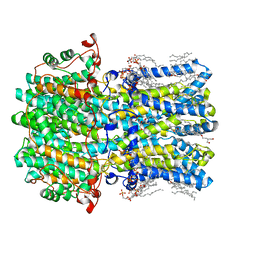 | | hBest2 Ca2+-unbound closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1M
 
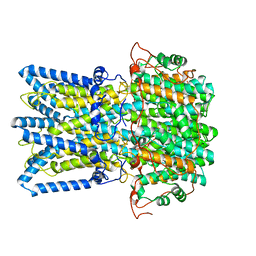 | |
3BY8
 
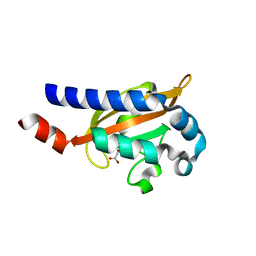 | | Crystal Structure of the E.coli DcuS Sensor Domain | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Sensor protein dcuS | | Authors: | Cheung, J, Hendrickson, W.A. | | Deposit date: | 2008-01-15 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of C4-Dicarboxylate Ligand Complexes with Sensor Domains of Histidine Kinases DcuS and DctB.
J.Biol.Chem., 283, 2008
|
|
6C5Y
 
 | | Crystal structure of thaumatin from microcrystals | | Descriptor: | Thaumatin-1 | | Authors: | Guo, G, Fuchs, M, Shi, W, Skinner, J, Berman, E, Ogata, C.M, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sample manipulation and data assembly for robust microcrystal synchrotron crystallography.
IUCrJ, 5, 2018
|
|
3BY9
 
 | |
3BQA
 
 | |
3BQ8
 
 | |
4I53
 
 | |
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JNF
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | Hsp70 CHAPERONE DnaK | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3I9Y
 
 | |
4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3H7M
 
 | |
3GTY
 
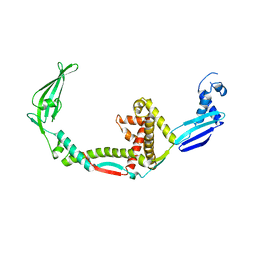 | |
3I9W
 
 | |
3GU0
 
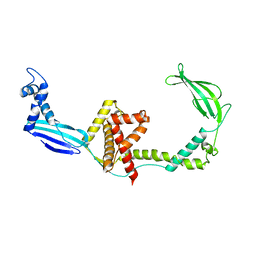 | |
4I54
 
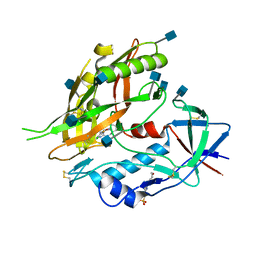 | |
1KPA
 
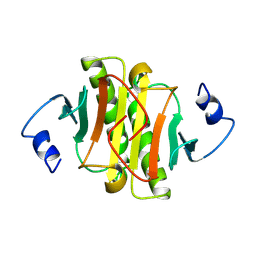 | | PKCI-1-ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1KPC
 
 | | PKCI-1-APO+ZINC | | Descriptor: | HUMAN PROTEIN KINASE C INTERACTING PROTEIN 1 (ZINC PROTEIN) | | Authors: | Lima, C.D, Klein, M.G, Weinstein, I.B, Hendrickson, W.A. | | Deposit date: | 1996-01-06 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of human protein kinase C interacting protein 1, a member of the HIT family of proteins.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
