4C4N
 
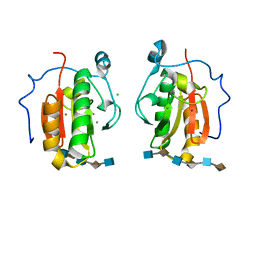 | | Crystal structure of the Sonic Hedgehog-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C4M
 
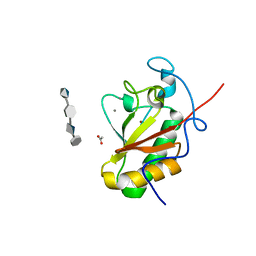 | | Crystal structure of the Sonic Hedgehog-chondroitin-4-sulphate complex | | Descriptor: | ACETATE ION, CALCIUM ION, SONIC HEDGEHOG PROTEIN, ... | | Authors: | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5EC5
 
 | | Crystal structure of lysenin pore | | Descriptor: | Lysenin, MERCURIBENZOIC ACID, MERCURY (II) ION | | Authors: | Podobnik, M, Savory, P, Rojko, N, Kisovec, M, Bruce, M, Jayasinghe, L, Anderluh, G. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of an invertebrate cytolysin pore reveals unique properties and mechanism of assembly.
Nat Commun, 7, 2016
|
|
3HHF
 
 | | Structure of CrgA regulatory domain, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
3HHG
 
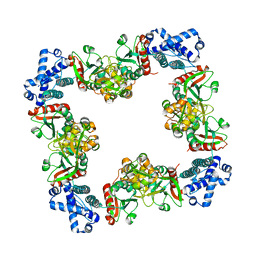 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
5J69
 
 | |
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
8A1S
 
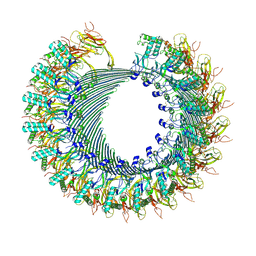 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
8A1D
 
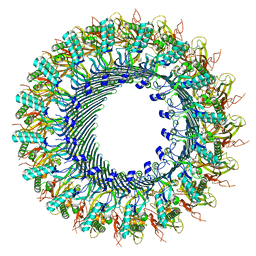 | | Structure of murine perforin-2 (Mpeg1) pore in ring form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
5J67
 
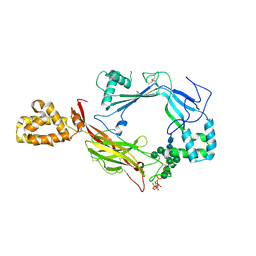 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
5J68
 
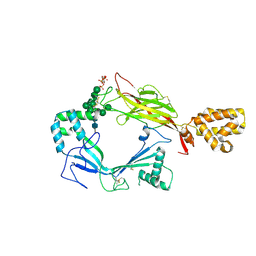 | | Structure of Astrotactin-2, a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Astrotactin-2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Ni, T, Harlos, K, Gilbert, R.J.C. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.221 Å) | | Cite: | Structure of astrotactin-2: a conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development.
Open Biology, 6, 2016
|
|
7ZMO
 
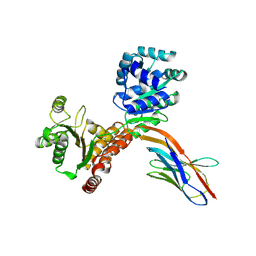 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-052 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-052, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMV
 
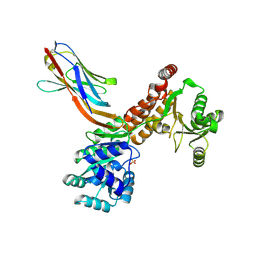 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMT
 
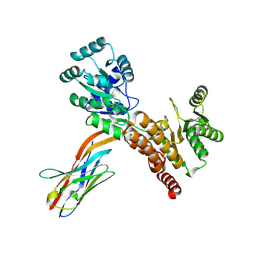 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMP
 
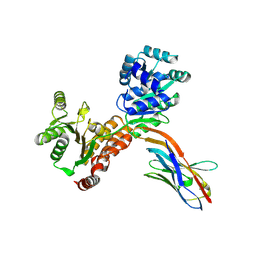 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMR
 
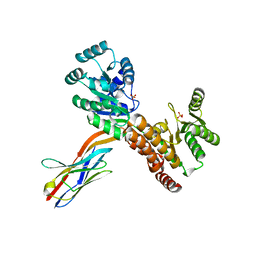 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-011 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-011, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMM
 
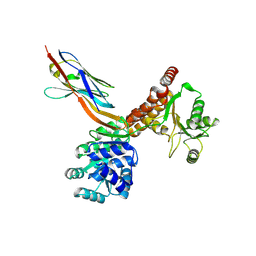 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2-001, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMN
 
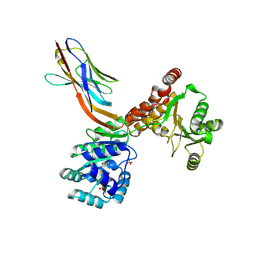 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-048 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-048, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMQ
 
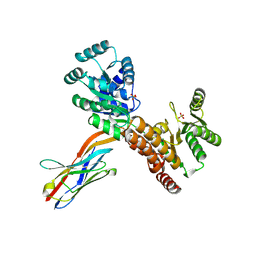 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZML
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G1-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G1-001, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMS
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G4-043 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G4-043, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
4C99
 
 | |
4C8F
 
 | | mouse ZNRF3 ectodomain crystal form IV | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C9A
 
 | |
4C8C
 
 | | mouse ZNRF3 ectodomain crystal form III | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
