2HII
 
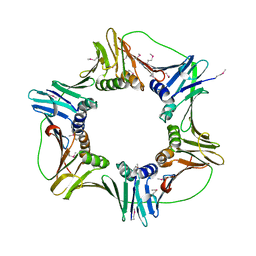 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HIK
 
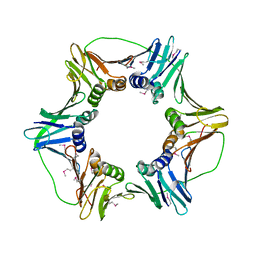 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
5E8G
 
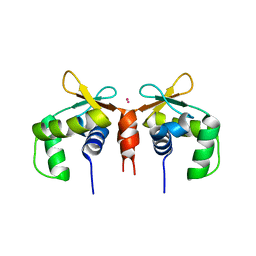 | |
5E8I
 
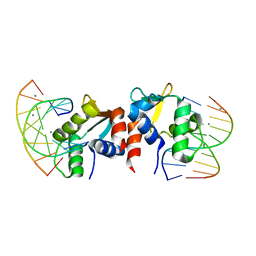 | |
9BE0
 
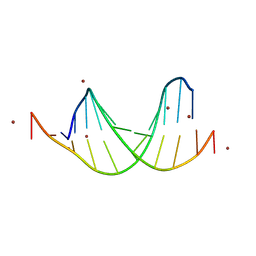 | | GC-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE1
 
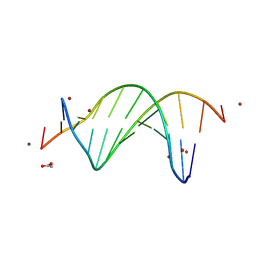 | | CG-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDZ
 
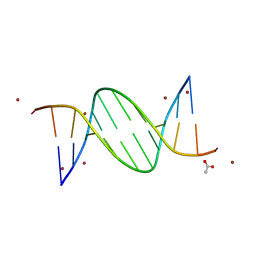 | | TA-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, ZINC ION, double-stranded DNA | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Wang, V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDY
 
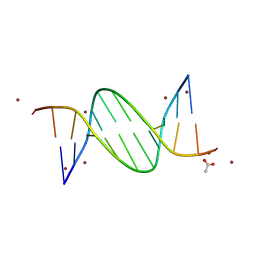 | | AT-centric NF-kappaB RelA binding DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2ONT
 
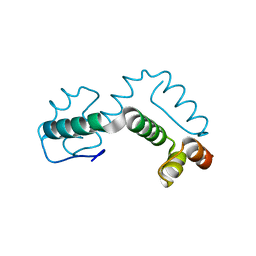 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5U
 
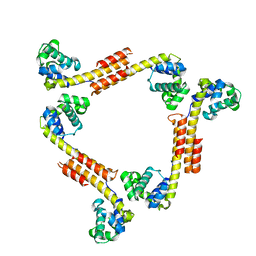 | |
4Z74
 
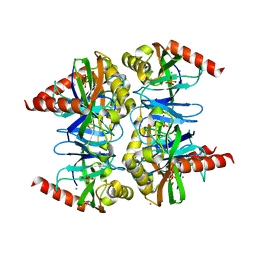 | |
4Z72
 
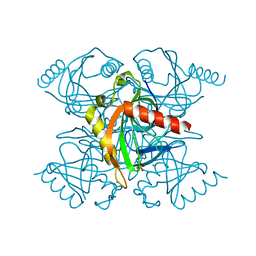 | | Crystal structure of inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with two phosphate ions | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PHOSPHATE ION | | Authors: | Pratt, A.C, Biswas, T, Barnard-Britson, S, Tsodikov, O.V. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural and computational dissection of the catalytic mechanism of the inorganic pyrophosphatase from Mycobacterium tuberculosis.
J.Struct.Biol., 192, 2015
|
|
4Z70
 
 | |
4Z73
 
 | | Crystal structure of inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with a phosphate ion and an inorganic pyrophosphate | | Descriptor: | Inorganic pyrophosphatase, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Pratt, A.C, Biswas, T, Tsodikov, O.V. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and computational dissection of the catalytic mechanism of the inorganic pyrophosphatase from Mycobacterium tuberculosis.
J.Struct.Biol., 192, 2015
|
|
4Z71
 
 | |
4RPA
 
 | |
4RV9
 
 | | Crystal structure of MtmC in complex with SAH | | Descriptor: | ACETATE ION, CHLORIDE ION, D-mycarose 3-C-methyltransferase, ... | | Authors: | Hou, C, Chen, J.-M, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2014-11-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
6OVQ
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | Descriptor: | GLYCEROL, Putative Side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OW0
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6VV0
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1354 | | Descriptor: | 2-[(4-amino-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(diethylamino)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VUY
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT358 | | Descriptor: | (7S)-7-phenyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VUZ
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT353 | | Descriptor: | 2-{[3-(piperidin-1-yl)propyl]sulfanyl}-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VUU
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1347 | | Descriptor: | (7S)-7-methyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-7,8-dihydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VV1
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT384 | | Descriptor: | 2-[(4-amino-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(piperidin-1-yl)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
