8EJE
 
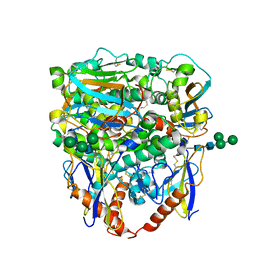 | | Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein GP1, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJF
 
 | | Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJH
 
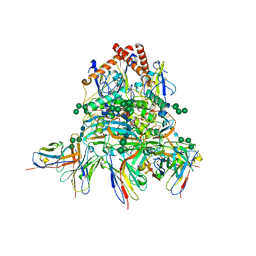 | | Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab | | Descriptor: | 12.1F Fab heavy chain, 12.1F Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
3SP7
 
 | | Crystal Structure of Bcl-xL bound to BM903 | | Descriptor: | 5-(4-chlorophenyl)-4-{3-[4-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]amino}phenyl)piperazin-1-yl]phenyl}-1,2-dimethyl-1H-pyrrole-3-carboxylic acid, ACETATE ION, Bcl-2-like protein 1, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of a new class of potent Bcl-2/Bcl-xL inhibitors
To be Published
|
|
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
7FCP
 
 | |
7FCQ
 
 | |
3QJZ
 
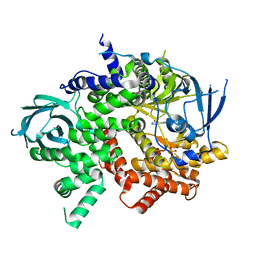 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
5RG4
 
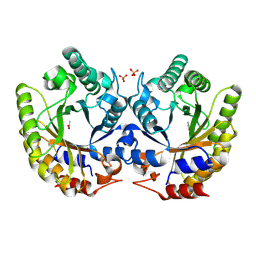 | | Crystal Structure of Kemp Eliminase HG3 in unbound state, 277K | | Descriptor: | ACETATE ION, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGA
 
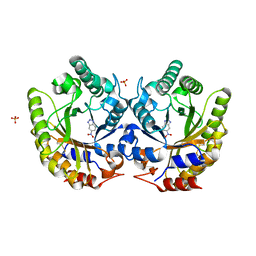 | | Crystal Structure of Kemp Eliminase HG3 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGC
 
 | | Crystal Structure of Kemp Eliminase HG3.7 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG7
 
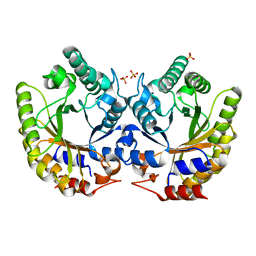 | | Crystal Structure of Kemp Eliminase HG3.14 in unbound state, 277K | | Descriptor: | Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
7W33
 
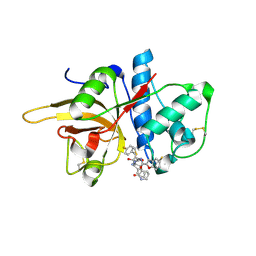 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
5HYX
 
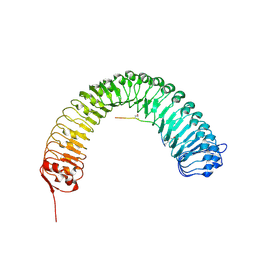 | | Plant peptide hormone receptor RGFR1 in complex with RGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-SER-ASN-PRO-GLY-HIS-HIS-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth
Cell Res., 26, 2016
|
|
7X7D
 
 | | SARS-CoV-2 Delta RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1 | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7X7E
 
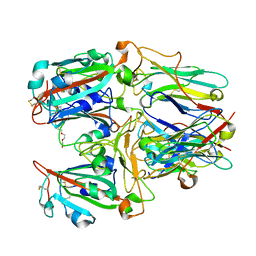 | | SARS-CoV-2 RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
5RGE
 
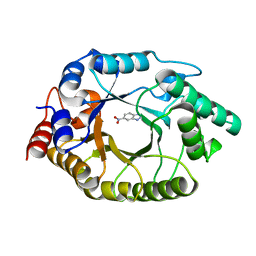 | | Crystal Structure of Kemp Eliminase HG3.17 with bound transition state analog, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3 | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RG9
 
 | | Crystal Structure of Kemp Eliminase HG4 in unbound state, 277K | | Descriptor: | ACETATE ION, Kemp Eliminase HG4, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RGF
 
 | | Crystal Structure of Kemp Eliminase HG4 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
6L0O
 
 | | WH domain of human MCM8 | | Descriptor: | DNA helicase MCM8, SULFATE ION | | Authors: | Liu, Y, Li, J, Liu, L, Zeng, H. | | Deposit date: | 2019-09-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of the winged-helix domain of MCM8.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
