3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
1UI8
 
 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
2ELG
 
 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
1V6C
 
 | | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11 | | Descriptor: | CALCIUM ION, SULFATE ION, alkaline serine protease, ... | | Authors: | Dong, D, Ihara, T, Motoshima, H, Watanabe, K. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11
To be Published
|
|
1V4P
 
 | |
1V2X
 
 | | TrmH | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (Gm18) methyltransferase | | Authors: | Nureki, O, Watanabe, K, Fukai, S, Ishii, R, Endo, Y, Hori, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deep Knot Structure for Construction of Active Site and Cofactor Binding Site of tRNA Modification Enzyme
STRUCTURE, 12, 2004
|
|
1Z9H
 
 | | Microsomal prostaglandin E synthase type-2 | | Descriptor: | ACETATE ION, CHLORIDE ION, INDOMETHACIN, ... | | Authors: | Yamada, T, Komoto, J, Watanabe, K, Ohmiya, Y, Takusagawa, F. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Possible Catalytic Mechanism of Microsomal Prostaglandin E Synthase Type 2 (mPGES-2).
J.Mol.Biol., 348, 2005
|
|
2E2T
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with phenylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E2U
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with 4-hydroxybenzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
1VC2
 
 | |
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2Z6O
 
 | | Crystal Structure of the Ufc1, Ufm1 conjugating enzyme 1 | | Descriptor: | MAGNESIUM ION, Ufm1-conjugating enzyme 1 | | Authors: | Mizushima , T, Tatsumi, K, Ozaki, Y, Kawakami, T, Suzuki, A, Ogasahara, K, Komatsu, M, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2007-08-06 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Ufc1, the Ufm1-conjugating enzyme
Biochem.Biophys.Res.Commun., 362, 2007
|
|
3A7S
 
 | | Catalytic domain of UCH37 | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Nishio, K, Kim, S.W, Kawai, K, Mizushima, T, Yamane, T, Hamazaki, J, Murata, S, Tanaka, K. | | Deposit date: | 2009-10-04 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain
Biochem.Biophys.Res.Commun., 2009
|
|
7DQE
 
 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7DQC
 
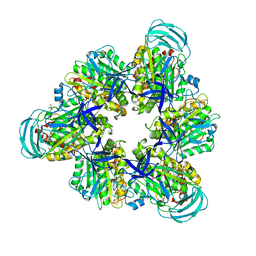 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
7EBC
 
 | | Crystal structure of Isocitrate lyase-1 from Saccaromyces cervisiae | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBF
 
 | | Cryo-EM structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | Isocitrate lyase | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
7EBE
 
 | | Crystal structure of Isocitrate lyase-1 from Candida albicans | | Descriptor: | FORMIC ACID, Isocitrate lyase, MAGNESIUM ION | | Authors: | Hiragi, K, Nishio, K, Moriyama, S, Hamaguchi, T, Mizoguchi, A, Yonekura, K, Tani, K, Mizushima, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the targeting specificity of ubiquitin ligase for S. cerevisiae isocitrate lyase but not C. albicans isocitrate lyase.
J.Struct.Biol., 213, 2021
|
|
2Z6P
 
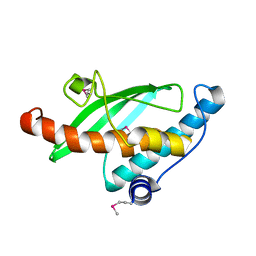 | | Crystal Structure of the Ufc1, Ufm1 conjugating enzyme 1 | | Descriptor: | Ufm1-conjugating enzyme 1 | | Authors: | Mizushima , T, Tatsumi, K, Ozaki, Y, Kawakami, T, Suzuki, A, Ogasahara, K, Komatsu, M, Kominami, E, Tanaka, K, Yamane, T. | | Deposit date: | 2007-08-06 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ufc1, the Ufm1-conjugating enzyme
Biochem.Biophys.Res.Commun., 362, 2007
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | Descriptor: | Claudin-19, Heat-labile enterotoxin B chain | | Authors: | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
6JPT
 
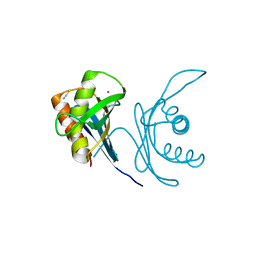 | | Crystal structure of human PAC3 homodimer (trigonal form) | | Descriptor: | POTASSIUM ION, Proteasome assembly chaperone 3, THIOCYANATE ION | | Authors: | Satoh, T, Yagi-Utsumi, M, Okamoto, K, Kurimoto, E, Tanaka, K, Kato, K. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Molecular and Structural Basis of the Proteasome alpha Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer.
Int J Mol Sci, 20, 2019
|
|
7CBD
 
 | | Catalytic domain of Cellulomonas fimi Cel6B | | Descriptor: | Exoglucanase A | | Authors: | Nakamura, A, Ishiwata, D, Visootsat, A, Uchiyama, T, Mizutani, K, Kaneko, S, Murata, T, Igarashi, K, Iino, R. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain architecture divergence leads to functional divergence in binding and catalytic domains of bacterial and fungal cellobiohydrolases.
J.Biol.Chem., 295, 2020
|
|
3VR2
 
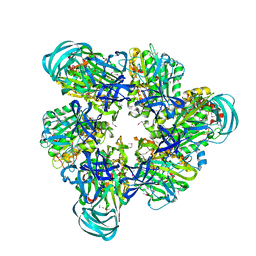 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3A2E
 
 | | Crystal structure of ginkbilobin-2, the novel antifungal protein from Ginkgo biloba seeds | | Descriptor: | Ginkbilobin-2 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases
Proteins, 77, 2009
|
|
