5EUF
 
 | | The crystal structure of a protease from Helicobacter pylori | | Descriptor: | GLYCEROL, Protease, ZINC ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a protease from Helicobacter pylori
To Be Published
|
|
5EZ4
 
 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5FD8
 
 | |
5END
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(Q152A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL | | Authors: | Hou, J, Cooper, D.R, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
5F9M
 
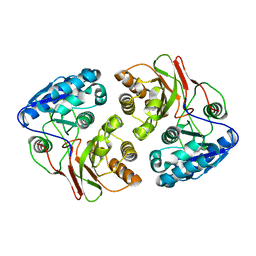 | | Crystal structure of native B3275, member of MccF family of enzymes | | Descriptor: | MccC family protein | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of native B3275, member of MccF family of enzymes.
To Be Published
|
|
5F7U
 
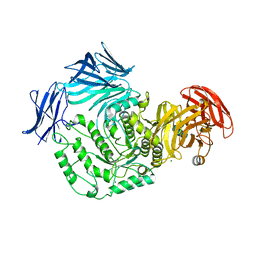 | |
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
5FCD
 
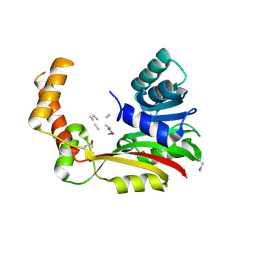 | | Crystal structure of MccD protein | | Descriptor: | CHLORIDE ION, MccD, UNK-UNK-UNK-MSE-UNK, ... | | Authors: | Nocek, B, Jedrzejczak, R, Anderson, W.F, Severinov, K, Dubiley, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MccD protein.
To Be Published
|
|
5HL3
 
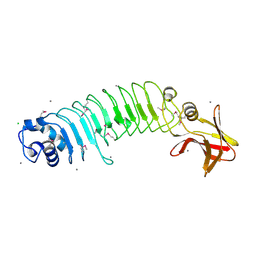 | | Crystal structure of Listeria monocytogenes InlP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lmo2470 protein | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Kiryukhina, O, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
5HNM
 
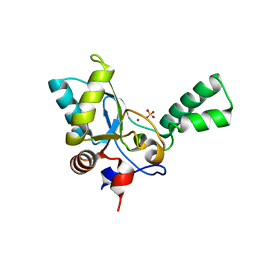 | | Crystal structure of vancomycin resistance D,D-pentapeptidase VanY E175A mutant from VanB-type resistance cassette in complex with Zn(II) | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, SULFATE ION, ZINC ION | | Authors: | Stogios, P.J, Chun, J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
5HPO
 
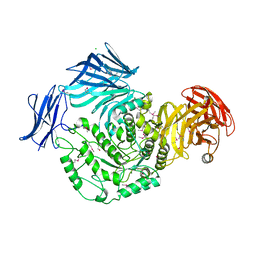 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5HT0
 
 | | Crystal structure of an Antibiotic_NAT family aminoglycoside acetyltransferase HMB0038 from an uncultured soil metagenomic sample in complex with coenzyme A | | Descriptor: | Aminoglycoside acetyltransferase HMB0005, COENZYME A, SULFATE ION | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
4E4Y
 
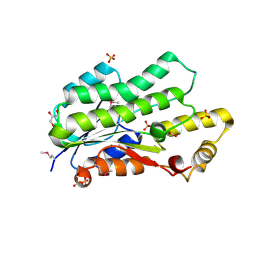 | | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | GLYCEROL, SULFATE ION, Short chain dehydrogenase family protein | | Authors: | Zhang, R, Zhou, M, Tan, K, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The crystal structure of a short chain dehydrogenase family protein from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4DQ6
 
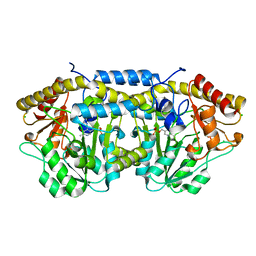 | | Crystal structure of PLP-bound putative aminotransferase from Clostridium difficile 630 | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Putative pyridoxal phosphate-dependent transferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Kudritska, M, Chruszcz, M, Grimshaw, S, Porebski, P.J, Cooper, D.R, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of putative aminotransferase from Clostridium difficile
630
to be published
|
|
4EP1
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Mikolajczak, K, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of anabolic ornithine carbamoyltransferase from Bacillus anthracis
To be Published
|
|
4E6Y
 
 | | Type II citrate synthase from Vibrio vulnificus. | | Descriptor: | Citrate synthase, FORMIC ACID | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Type II citrate synthase from Vibrio vulnificus.
To be Published
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4EAE
 
 | | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e | | Descriptor: | D-MALATE, Lmo1068 protein, SODIUM ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e
To be Published
|
|
4ECL
 
 | | Crystal structure of the cytoplasmic domain of vancomycin resistance serine racemase VanTg | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine racemase | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Cosme, J, Di Leo, R, Krishnamoorthy, M, Meziane-Cherif, D, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structural and Functional Adaptation of Vancomycin Resistance VanT Serine Racemases.
MBio, 6, 2015
|
|
4EGU
 
 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
4E4R
 
 | | EutD phosphotransacetylase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | EutD phosphotransacetylase from Staphylococcus aureus.
To be Published
|
|
4EWQ
 
 | | Human p38 alpha MAPK in complex with a pyridazine based inhibitor | | Descriptor: | 3-phenyl-4-(pyridin-4-yl)-6-[4-(pyrimidin-2-yl)piperazin-1-yl]pyridazine, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ACETATE ION, ... | | Authors: | Grum-Tokars, V, Minasov, G, Roy, S, Schavocky, J, Winsor, J, Watterson, D.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
4DFB
 
 | | Crystal structure of aminoglycoside phosphotransferase aph(2")-id/aph(2")-iva in complex with kanamycin | | Descriptor: | APH(2")-Id, CHLORIDE ION, KANAMYCIN A | | Authors: | Stogios, P.J, Minasov, G, Osipiuk, J, Evdokimova, E, Egorova, E, Di leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
