7LVM
 
 | |
7LVJ
 
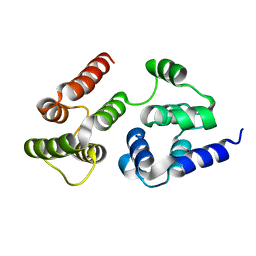 | | CASP8 isoform G DED domain | | Descriptor: | Isoform 9 of Caspase-8 | | Authors: | Weichert, K, Lu, F, Kodandapani, L, Sauder, J.M. | | Deposit date: | 2021-02-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caspase-8 Variant G Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Aggressive Behavior.
ACR Open Rheumatol, 4, 2022
|
|
4QZT
 
 | |
4R0W
 
 | |
8IJ1
 
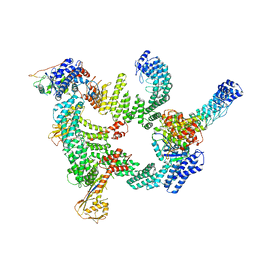 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
5ZIT
 
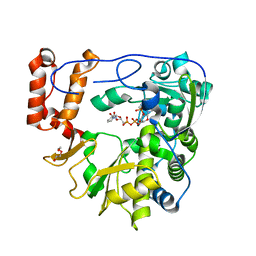 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
5X6O
 
 | | Intact ATR/Mec1-ATRIP/Ddc2 complex | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Wang, X, Ran, T, Cai, G. | | Deposit date: | 2017-02-22 | | Release date: | 2017-12-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Science, 358, 2017
|
|
7KZY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM778 (3-methyl-N-(1-(5-methyl-1H-pyrazol-3-yl)ethyl)-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1H-pyrazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7L0K
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM589 (ethyl 3-methyl-4-((4-(trifluoromethyl)benzo[d]oxazol-7-yl)methyl)-1H-pyrrole-2-carboxylate) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZ4
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM705 (N-(1-(1H-1,2,4-triazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(1H-1,2,4-triazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7MK1
 
 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
8JE2
 
 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | Descriptor: | Cullin-2, Elongin-B, Elongin-C, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
8JE1
 
 | | An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
7D0J
 
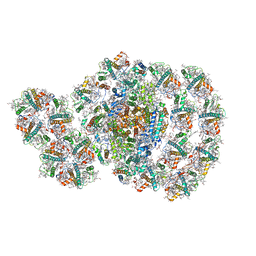 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
4R0U
 
 | |
7N1R
 
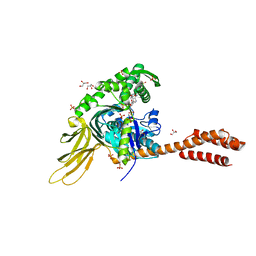 | | A novel and unique ATP hydrolysis to AMP by a human Hsp70 BiP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Yang, J, Musayev, F, Liu, Q. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).
Protein Sci., 31, 2022
|
|
5YUD
 
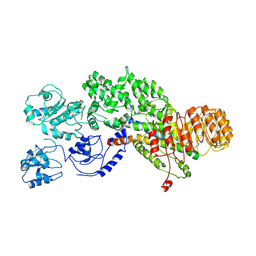 | | Flagellin derivative in complex with the NLR protein NAIP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | Authors: | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
4ID1
 
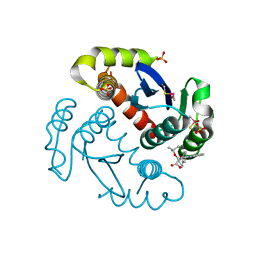 | |
4LNU
 
 | | Nucleotide-free kinesin motor domain in complex with tubulin and a DARPin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Designed ankyrin repeat protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Cao, L, Gigant, B, Knossow, M. | | Deposit date: | 2013-07-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement
Nat Commun, 5, 2014
|
|
6W3H
 
 | | Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATV Fc, Transferrin receptor protein 1,Transferrin receptor protein 1 | | Authors: | Srivastava, A, Kariolis, M, Wells, R. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys.
Sci Transl Med, 12, 2020
|
|
7BR3
 
 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
6N4G
 
 | |
2FSA
 
 | |
2FUI
 
 | |
