7YR9
 
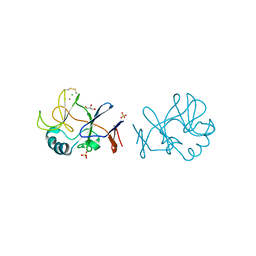 | | Crystal structure of the immature form of TtPetA | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tsutsumi, E, Niwa, S, Takeda, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.
Commun Chem, 6, 2023
|
|
3VOA
 
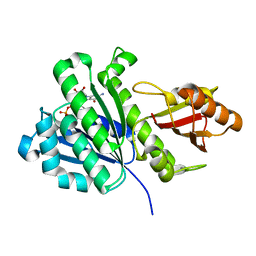 | | Staphylococcus aureus FtsZ 12-316 GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VO9
 
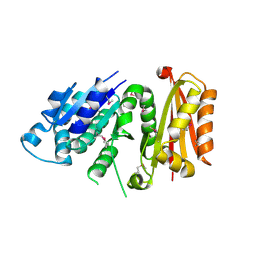 | | Staphylococcus aureus FtsZ apo-form (SeMet) | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3CLE
 
 | |
3VO8
 
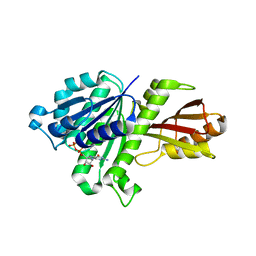 | | Staphylococcus aureus FtsZ GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VPA
 
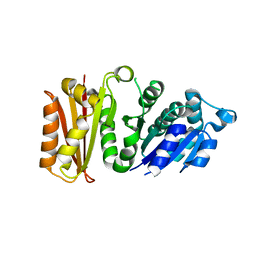 | | Staphylococcus aureus FtsZ apo-form | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7D69
 
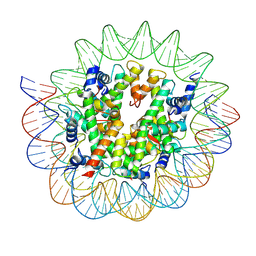 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
3VJI
 
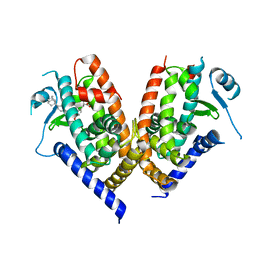 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
4G6F
 
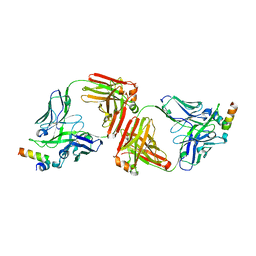 | | Crystal Structure of 10E8 Fab in Complex with an HIV-1 gp41 Peptide | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, gp41 MPER Peptide | | Authors: | Ofek, G, Huang, J, Connors, M, Kwong, P.D. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad and potent neutralization of HIV-1 by a gp41-specific human antibody.
Nature, 491, 2012
|
|
8YTN
 
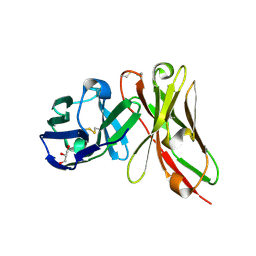 | | Single-chain Fv antibody of E11 | | Descriptor: | GLYCEROL, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YY8
 
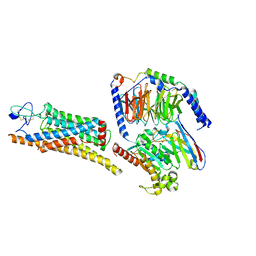 | | Fzd7 -Gs complex | | Descriptor: | Frizzled-7, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B, Xu, L, Han, G.W, Xu, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structure of constitutively active human Frizzled 7 in complex with heterotrimeric G s .
Cell Res., 31, 2021
|
|
8YTP
 
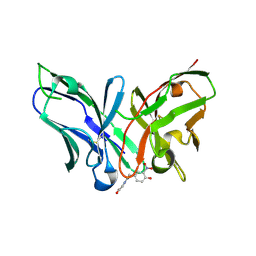 | | Single-chain Fv antibody of E11 complex with NP-glycine under reducing conditions | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTO
 
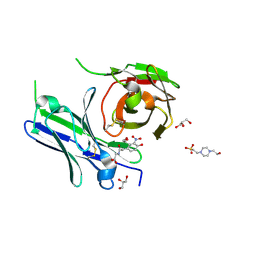 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
7ZJQ
 
 | |
1GSA
 
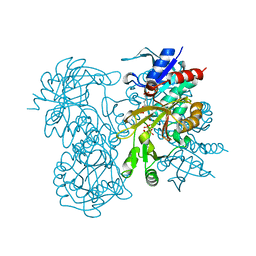 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
7DB6
 
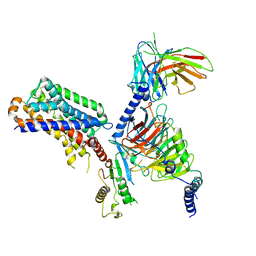 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6L4Q
 
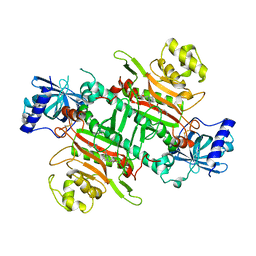 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-B | | Descriptor: | (3R)-3-[[(3R)-3-methylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y, Mishra, S, Harlos, K. | | Deposit date: | 2019-10-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
6L3Y
 
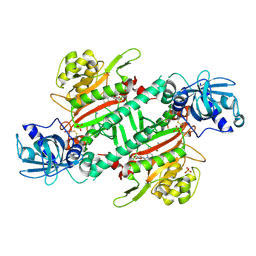 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-C | | Descriptor: | (3R)-3-[[(3S)-3-ethylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Babbar, P, Sharma, A, Mishra, S, Manickam, Y, Harlos, K. | | Deposit date: | 2019-10-15 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
1OCR
 
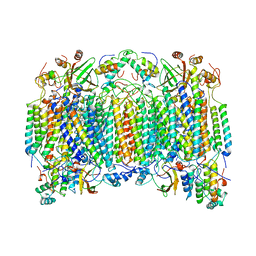 | |
1OCO
 
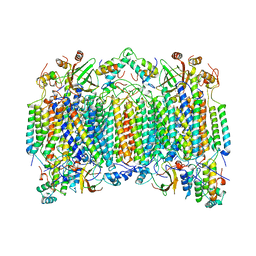 | |
1OCZ
 
 | |
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1GDE
 
 | | CRYSTAL STRUCTURE OF PYROCOCCUS PROTEIN A-1 E-FORM | | Descriptor: | ASPARTATE AMINOTRANSFERASE, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-23 | | Release date: | 2001-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1HMK
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
1G5P
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Strop, P, Takahara, P.M, Chiu, H.J, Angove, H.C, Burgess, B.K, Rees, D.C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the all-ferrous [4Fe-4S]0 form of the nitrogenase iron protein from Azotobacter vinelandii.
Biochemistry, 40, 2001
|
|
