7MWI
 
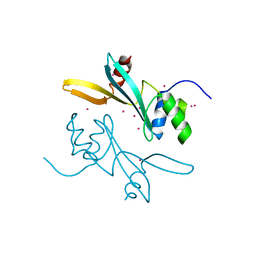 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
5ZHG
 
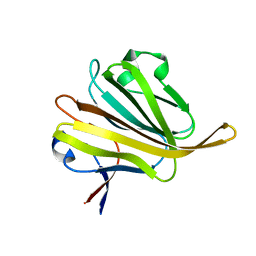 | |
4TZU
 
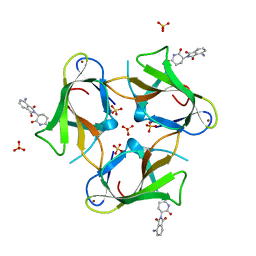 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | Descriptor: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4TZC
 
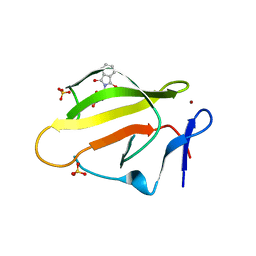 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
7BPC
 
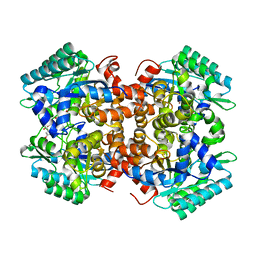 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BP1
 
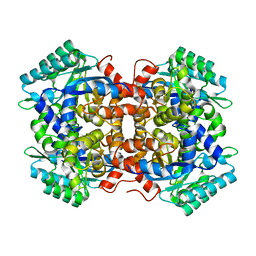 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
5ZHO
 
 | |
8WOU
 
 | |
8WKJ
 
 | |
5PGY
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
4U93
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
5PGZ
 
 | | CRYSTAL STRUCTURE OF MURINE 11BETA- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE (BMS-816336) | | Descriptor: | 2-[(5R,7S)-6-HYDROXY-2-PHENYLADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
7TTM
 
 | |
7TTX
 
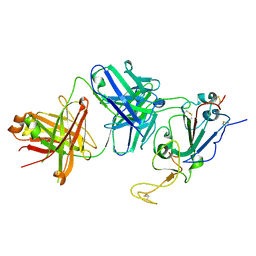 | |
7YV4
 
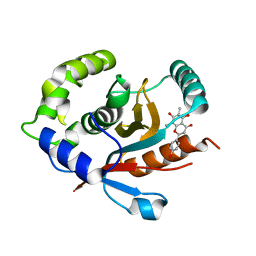 | | Crystal structure of human UCHL3 in complex with Farrerol | | Descriptor: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
7TTY
 
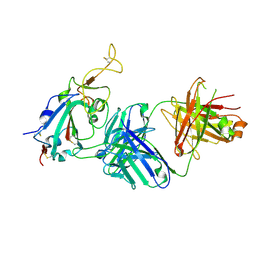 | |
6KUR
 
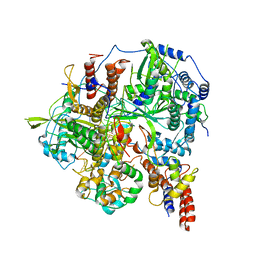 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
6KUV
 
 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5PGU
 
 | |
6KV5
 
 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
