7UXJ
 
 | | Structure of PPIA in complex with FP29102, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP29102, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYJ
 
 | | Structure of RNF31 in complex with FP06652, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06652, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXN
 
 | | Structure of PPIA in complex with FP29103, a Helicon Polypeptide | | Descriptor: | FP29103, N,N'-(1,4-phenylene)diacetamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6X4A
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | Descriptor: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
6IJT
 
 | |
6JYS
 
 | |
6JYO
 
 | |
6JYR
 
 | |
6JYN
 
 | |
6MVH
 
 | |
6MVF
 
 | |
1LPF
 
 | |
2GZQ
 
 | |
2FVC
 
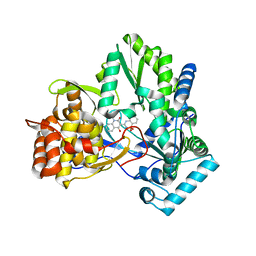 | | Crystal structure of NS5B BK strain (delta 24) in complex with a 3-(1,1-Dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinone | | Descriptor: | 3-(1,1-dioxido-4H-1,2,4-benzothiadiazin-3-yl)-4-hydroxy-1-(3-methylbutyl)quinolin-2(1H)-one, polyprotein | | Authors: | Concha, N.O, Wonacott, A, Singh, O. | | Deposit date: | 2006-01-30 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase.
J.Med.Chem., 49, 2006
|
|
1PHH
 
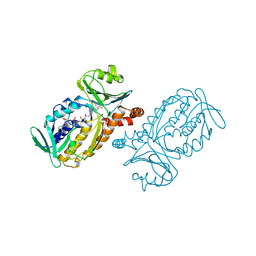 | | CRYSTAL STRUCTURE OF P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH ITS REACTION PRODUCT 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Drenth, J. | | Deposit date: | 1987-11-04 | | Release date: | 1988-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of p-hydroxybenzoate hydroxylase complexed with its reaction product 3,4-dihydroxybenzoate.
J.Mol.Biol., 199, 1988
|
|
2F8M
 
 | |
2JNR
 
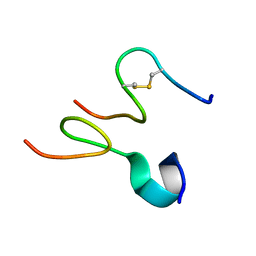 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
2L6S
 
 | | Efficacy of an HIV-1 entry inhibitor targeting the GP41 fusion peptide | | Descriptor: | VIR-576 | | Authors: | Forssmann, W, The, Y, Stoll, M, Adermann, K, Albrecht, U, Barlos, K, Busmann, A, Canales-Mayordomo, A, Gimenez-Gallego, G, Hirsch, J, Jimenez-Barbero, J, Meyer-Olson, D, Muench, J, Perez-Castells, J, Standker, L, Kirchhoff, F, Schmidt, R.E. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide.
Sci Transl Med, 2, 2010
|
|
2LVJ
 
 | | solution structure of hemi-Mg-bound Phl p 7 | | Descriptor: | MAGNESIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
2LVI
 
 | | Solution structure of apo-Phl p 7 | | Descriptor: | Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
2LVK
 
 | | Solution structure of Ca-bound Phl p 7 | | Descriptor: | CALCIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Sirianni, A.G, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
4DNI
 
 | | Structure of Editosome protein | | Descriptor: | Fusion protein of RNA-editing complex proteins MP42 and MP18 | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-08 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.
J.Struct.Biol., 180, 2012
|
|
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
2N0X
 
 | | Three dimensional structure of EPI-X4, a human albumin-derived peptide that regulates innate immunity through the CXCR4/CXCL12 chemotactic axis and antagonizes HIV-1 entry | | Descriptor: | Serum albumin | | Authors: | Perez-Castells, J, Canales, A, Jimenez-Barbero, J, Gimenez-Gallego, G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and characterization of an endogenous CXCR4 antagonist.
Cell Rep, 11, 2015
|
|
