3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
3H0P
 
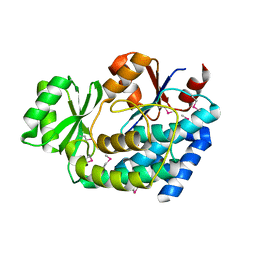 | | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium. | | Descriptor: | S-malonyltransferase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3H5Q
 
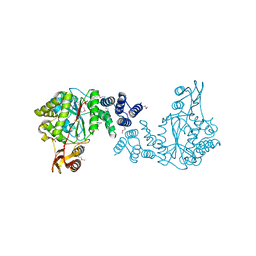 | | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SULFATE ION, THYMIDINE | | Authors: | Shumilin, I.A, Zimmerman, M, Cymborowski, M, Skarina, T, Onopriyenko, O, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3H2Y
 
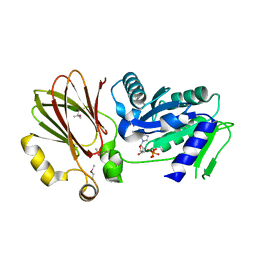 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
3HFR
 
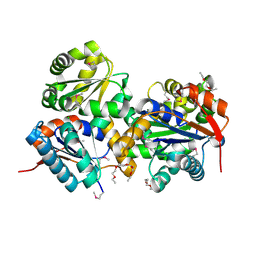 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
3I3W
 
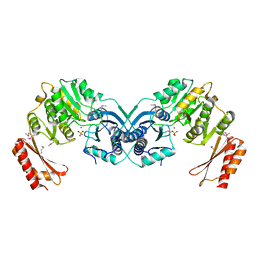 | | Structure of a phosphoglucosamine mutase from Francisella tularensis | | Descriptor: | Phosphoglucosamine mutase, ZINC ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a phosphoglucosamine mutase from Francisella tularensis
To be Published
|
|
4I62
 
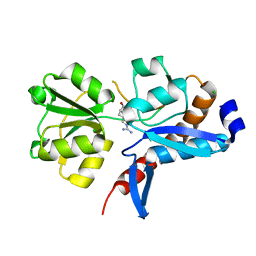 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
3T32
 
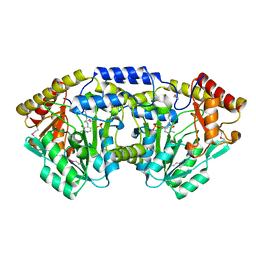 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
4MUQ
 
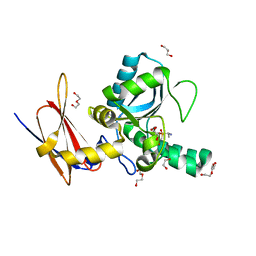 | | Crystal Structure of Vancomycin Resistance D,D-dipeptidase VanXYg in complex with D-Ala-D-Ala phosphinate analog | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, (2R)-3-[(R)-[(1S)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.364 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MUR
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3IMF
 
 | | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3IAC
 
 | | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium. | | Descriptor: | CHLORIDE ION, Glucuronate isomerase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium.
To be Published
|
|
3IHS
 
 | | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Brunzelle, J.S, Anderson, S.M, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames
To be Published
|
|
3SKS
 
 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
8SDC
 
 | | Crystal structure of fluoroacetate dehalogenase Daro3835 apoenzyme | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
4H59
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane
To be Published
|
|
3GEU
 
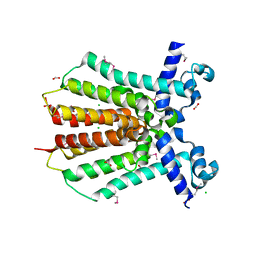 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
4NVR
 
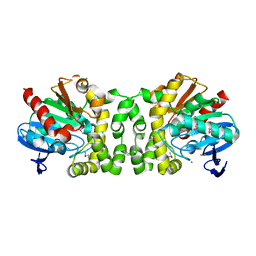 | | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Stam, J, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.22 Angstrom Resolution Crystal Structure of a Putative Acyltransferase from Salmonella enterica.
TO BE PUBLISHED
|
|
3T7Y
 
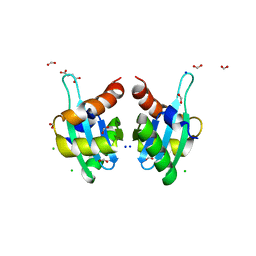 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
3T5P
 
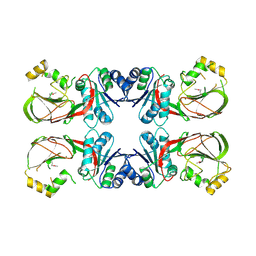 | | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne | | Descriptor: | BmrU protein, MAGNESIUM ION | | Authors: | Hou, J, Zheng, H, Chruszcz, M, Cooper, D.R, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative diacylglycerol kinase from Bacillus anthracis str. Sterne
To be Published
|
|
4GUC
 
 | | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein BA_2500, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames
To be Published
|
|
4JG9
 
 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
3GE1
 
 | | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Brunzelle, J, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol
To be Published
|
|
4N3O
 
 | | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni. | | Descriptor: | CALCIUM ION, Putative D-glycero-D-manno-heptose 7-phosphate kinase | | Authors: | Minasov, G, Wawrzak, Z, Gordon, E, Onopriyenko, O, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Resolution Crystal Structure of Putative Sugar Kinase from Campylobacter jejuni.
TO BE PUBLISHED
|
|
