7RXL
 
 | |
5TGV
 
 | |
5U7G
 
 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U64
 
 | | Camel nanobody VHH-28 | | Descriptor: | VHH-28 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
5UF1
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 in complex with H-trisaccharide | | Descriptor: | CHLORIDE ION, O13, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5U36
 
 | | Crystal Structure Of A Mutant M. Jannashii Tyrosyl-tRNA Synthetase | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Luo, X, Fu, G, Zhu, X, Wilson, I.A, Wang, F. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria.
Nat. Chem. Biol., 13, 2017
|
|
5UFC
 
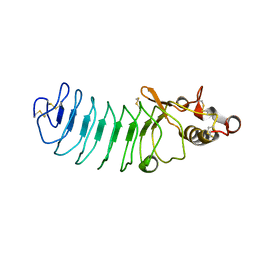 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 with H-trisaccharide bound | | Descriptor: | Tn4-22, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UI4
 
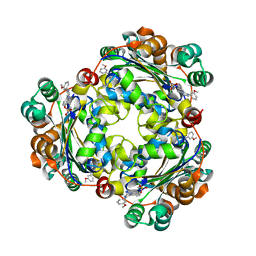 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | Descriptor: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | Authors: | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
5UXQ
 
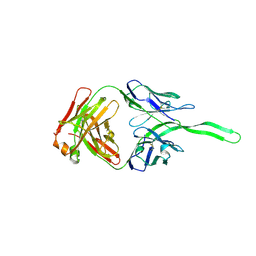 | | Structure of anti-HIV trimer apex antibody PGT143 | | Descriptor: | PGT143 Fab Heavy Chain, PGT143 Fab Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic Beta-Hairpin Structure
Immunity, 46, 2017
|
|
5UY3
 
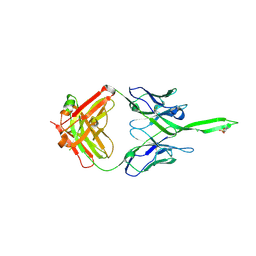 | | Crystal structure of human Fab PGT144, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Antibody PGT144 Fab heavy chain, Antibody PGT144 Fab light chain | | Authors: | Julien, J.-P, Lee, J.H, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
5V7R
 
 | |
5W3Q
 
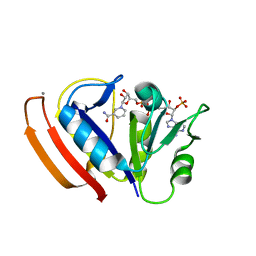 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
5VLI
 
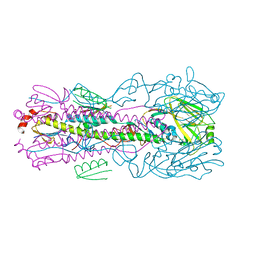 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
5VJL
 
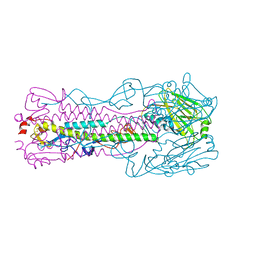 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
5VTW
 
 | |
5VTV
 
 | |
5VTQ
 
 | |
5VTX
 
 | |
5VTY
 
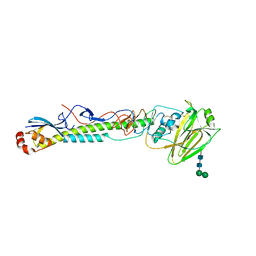 | |
5UFB
 
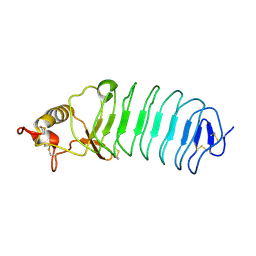 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 (Apo) | | Descriptor: | Tn4-22 | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFF
 
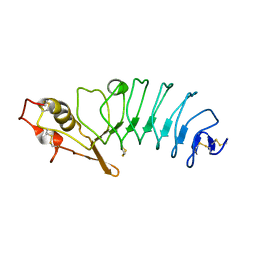 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 with Fucose(alpha-1-2)Lactose bound | | Descriptor: | RBC36, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5VTU
 
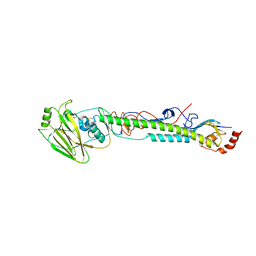 | |
5VTR
 
 | |
5VJK
 
 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
5VU4
 
 | |
