2YSR
 
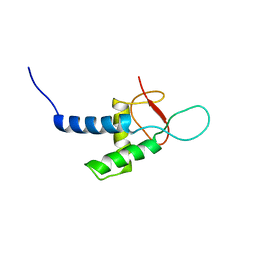 | |
2YTI
 
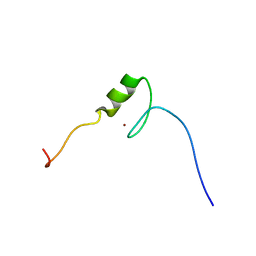 | | Solution structure of the C2H2 type zinc finger (region 564-596) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 564-596) of human Zinc finger protein 347
To be Published
|
|
2YTU
 
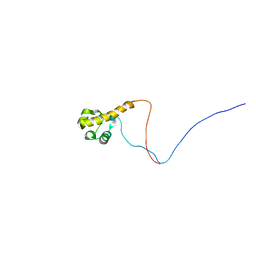 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A.K, Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
2YUK
 
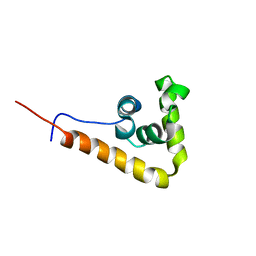 | | Solution structure of the HMG box of human Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog | | Descriptor: | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box of human Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog
To be Published
|
|
2YUV
 
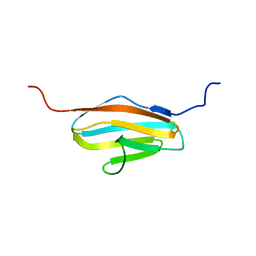 | | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C
To be Published
|
|
2YQE
 
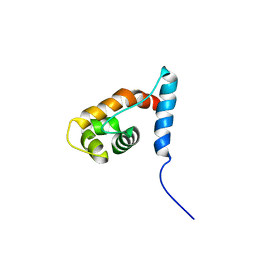 | | Solution structure of the ARID domain of JARID1D protein | | Descriptor: | Jumonji/ARID domain-containing protein 1D | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ARID domain of JARID1D protein
To be Published
|
|
2YRE
 
 | |
2YRY
 
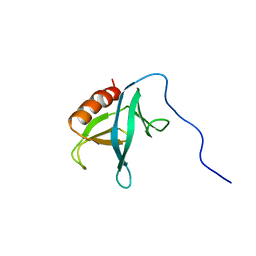 | | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human | | Descriptor: | Pleckstrin homology domain-containing family A member 6 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human
To be Published
|
|
2YWV
 
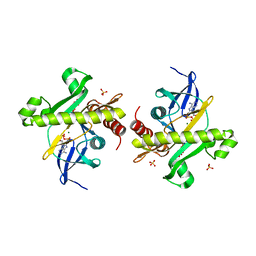 | | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole succinocarboxamide synthetase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of SAICAR synthetase from Geobacillus kaustophilus
To be Published
|
|
2YSJ
 
 | |
2YT7
 
 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3
To be Published
|
|
2YTR
 
 | | Solution structure of the C2H2 type zinc finger (region 760-792) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 760-792) of human Zinc finger protein 347
To be Published
|
|
2YXG
 
 | |
2YYE
 
 | | Crystal structure of selenophosphate synthetase from Aquifex aeolicus complexed with AMPCPP | | Descriptor: | COBALT (II) ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PHOSPHATE ION, ... | | Authors: | Itoh, Y, Sekine, S, Matsumoto, E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-29 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of selenophosphate synthetase essential for selenium incorporation into proteins and RNAs
J.Mol.Biol., 385, 2009
|
|
2YZR
 
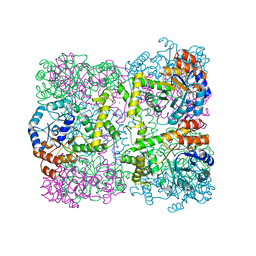 | |
2Z0B
 
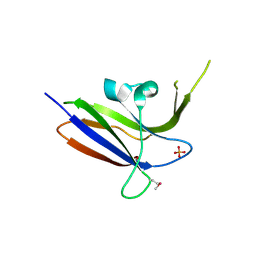 | | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434) | | Descriptor: | PHOSPHATE ION, Putative glycerophosphodiester phosphodiesterase 5 | | Authors: | Saijo, S, Nishino, A, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434)
To be Published
|
|
2Z0P
 
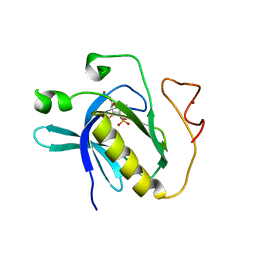 | | Crystal structure of PH domain of Bruton's tyrosine kinase | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Tyrosine-protein kinase BTK, ZINC ION | | Authors: | Murayama, K, Kato-Murayama, M, Mishima, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of the Bruton's tyrosine kinase PH domain with phosphatidylinositol
Biochem.Biophys.Res.Commun., 377, 2008
|
|
2YU8
 
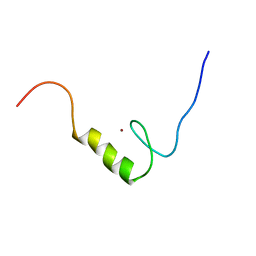 | | Solution structure of the C2H2 type zinc finger (region 648-680) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 648-680) of human Zinc finger protein 347
To be Published
|
|
2YUF
 
 | |
2YUX
 
 | | Solution Structure of 3rd Fibronectin type three Domain of slow type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 3rd Fibronectin type three Domain of slow type Myosin-Binding Protein C
To be Published
|
|
2YW9
 
 | |
2YXS
 
 | | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose | | Descriptor: | Galectin-8 variant, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose
To be Published
|
|
2YY3
 
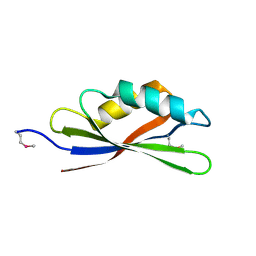 | |
2YQM
 
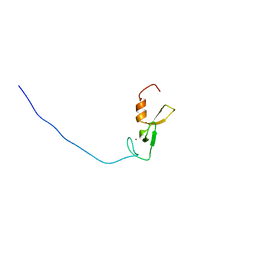 | | Solution structure of the FYVE domain in zinc finger FYVE domain-containing protein 12 | | Descriptor: | RUN and FYVE domain-containing protein 1, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the FYVE domain in zinc finger FYVE domain-containing protein 12
To be Published
|
|
2YRM
 
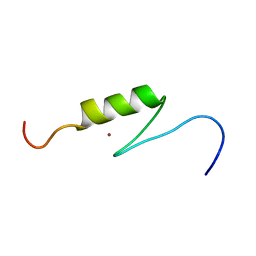 | | Solution structure of the 1st zf-C2H2 domain from Human B-cell lymphoma 6 protein | | Descriptor: | B-cell lymphoma 6 protein, ZINC ION | | Authors: | Tomizawa, T, Saito, K, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 1st zf-C2H2 domain from Human B-cell lymphoma 6 protein
To be Published
|
|
