2O3J
 
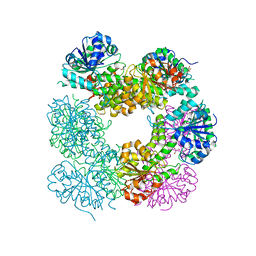 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
2PPG
 
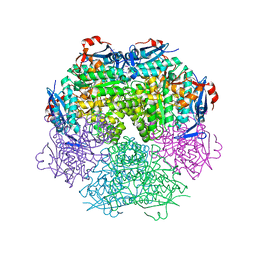 | | Crystal structure of putative isomerase from Sinorhizobium meliloti | | Descriptor: | Putative isomerase | | Authors: | Ramagopal, U.A, Toro, R, Dickey, M, Logan, C, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative isomerase from Sinorhizobium meliloti.
To be Published
|
|
2NS9
 
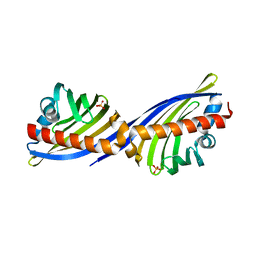 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
3NEK
 
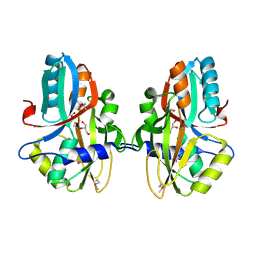 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
3H14
 
 | | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, classes I and II, GLYCEROL | | Authors: | Sampathkumar, P, Atwell, S, Wasserman, S, Miller, S, Bain, K, Rutter, M, Tarun, G, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-05-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative aminotransferase from Silicibacter pomeroyi
TO BE PUBLISHED
|
|
3HG7
 
 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
2OQH
 
 | |
2OO3
 
 | | Crystal structure of protein LPL1258 from Legionella pneumophila str. Philadelphia 1, Pfam DUF519 | | Descriptor: | Protein involved in catabolism of external DNA, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Slocombe, A, Reyes, C, Ozyurt, S, Smyth, L, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hypothetical lpl1258 protein from Legionella pneumophila
To be Published
|
|
2OY9
 
 | |
2OYN
 
 | | Crystal structure of CDP-bound protein MJ0056 from Methanococcus jannaschii, Pfam DUF120 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, Hypothetical protein MJ0056, SODIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of hypothetical protein from Methanococcus jannaschii bound to CDP
TO BE PUBLISHED
|
|
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
2PCE
 
 | | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | PHOSPHATE ION, putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM
To be Published
|
|
2P76
 
 | | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus
To be Published
|
|
2P61
 
 | | Crystal structure of protein TM1646 from Thermotoga maritima, Pfam DUF327 | | Descriptor: | Hypothetical protein TM_1646 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein TM_1646 from Thermotoga maritima
To be Published
|
|
2PAG
 
 | | Crystal structure of protein PSPTO_5518 from Pseudomonas syringae pv. tomato | | Descriptor: | CALCIUM ION, Hypothetical protein | | Authors: | Fedorov, A.A, Ramagopal, U, Toro, R, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Pseudomonas syringae pv. tomato.
To be Published
|
|
2PBZ
 
 | |
2OO2
 
 | | Crystal structure of protein AF1782 from Archaeoglobus fulgidus, Pfam DUF357 | | Descriptor: | Hypothetical protein AF_1782 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Adams, J, Sridhar, V, Smyth, L, Freeman, J, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical AF_1782 protein from Archaeoglobus fulgidus
To be Published
|
|
2OVL
 
 | |
3EZY
 
 | | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Dehydrogenase | | Authors: | Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Maletic, M, Gheyi, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-24 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima
To be published
|
|
3KKE
 
 | | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, LacI family Transcriptional regulator | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Do, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis
To be Published
|
|
3KGZ
 
 | | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris | | Descriptor: | Cupin 2 conserved barrel domain protein, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris
To be Published
|
|
3F1C
 
 | | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes | | Descriptor: | Putative 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase 2 | | Authors: | Patskovsky, Y, Ho, J, Toro, R, Gilmore, M, Miller, S, Groshong, C, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes
To be Published
|
|
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
3H7V
 
 | | CRYSTAL STRUCTURE OF O-SUCCINYLBENZOATE SYNTHASE FROM THERMOSYNECHOCOCCUS ELONGATUS BP-1 complexed with MG in the active site | | Descriptor: | MAGNESIUM ION, O-SUCCINYLBENZOATE SYNTHASE | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3L0Q
 
 | |
