7LZT
 
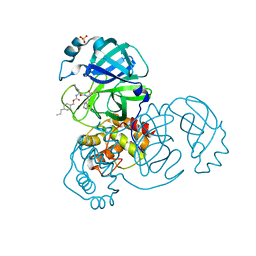 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8b | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)methoxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZX
 
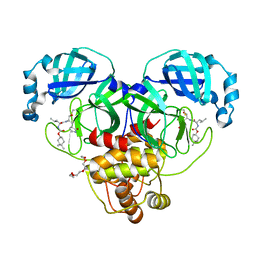 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 1c | | Descriptor: | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7LZU
 
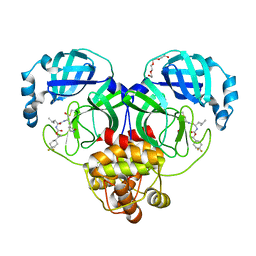 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 12b | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7M04
 
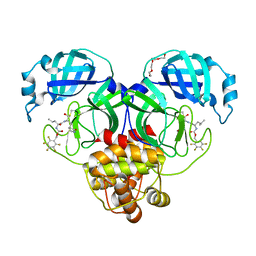 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 21c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7UHJ
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (60 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHP
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (2000 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHN
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (300 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHT
 
 | | SSX Structure of Metallo Beta-Lactamase L1 with One Zinc in the Active Site | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHK
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (80 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHH
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (20 ms snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHL
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (100 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHI
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (40 ms Snapshot) | | Descriptor: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHR
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 Before Reaction (Dark-Set) | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHQ
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (4000 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHS
 
 | | SSX Structure of Metallo Beta-Lactamase L1 with Two Water Molecules in the Active Site | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7K0E
 
 | | 1.90 A resolution structure of SARS-CoV-2 3CL protease in complex with deuterated GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-09-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0G
 
 | | 1.85 A resolution structure of SARS-CoV 3CL protease in complex with deuterated GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-09-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0H
 
 | | 1.70 A resolution structure of SARS-CoV 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | Descriptor: | CHLORIDE ION, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, Replicase polyprotein 1a, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-09-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7UH4
 
 | | LXG-associated alpha-helical protein D2 (LapD2) | | Descriptor: | LXG-associated alpha-helical protein D2, SULFATE ION | | Authors: | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
7UHM
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (150 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHO
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (500 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7LDQ
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846 | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-13 | | Release date: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846
To Be Published
|
|
3T1I
 
 | | Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Double-strand break repair protein MRE11A, GLYCEROL, ... | | Authors: | Park, Y.B, Chae, J, Kim, Y, Cho, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human mre11: understanding tumorigenic mutations
Structure, 19, 2011
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
5DGJ
 
 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
