1IYE
 
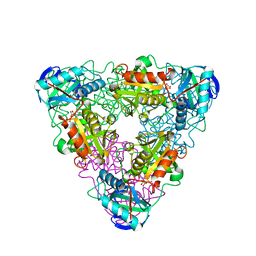 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Hirotsu, K, Goto, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of branched-chain amino Acid aminotransferase complexed with glutamate and glutarate: true reaction intermediate and double substrate recognition of the enzyme.
Biochemistry, 42, 2003
|
|
6LUT
 
 | |
8HR3
 
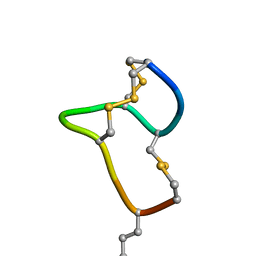 | | [D-Cys5,Asp7,Val8,D-Lys16]-STp(5-17) | | Descriptor: | DCY-CYS-ASP-VAL-CYS-CYS-ASN-PRO-ALA-CYS-ALA-DLY-CYS | | Authors: | Shimamoto, S, Hidaka, Y, Yoshino, S, Goto, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Molecular Basis of Heat-Stable Enterotoxin for Vaccine Development and Cancer Cell Detection.
Molecules, 28, 2023
|
|
1IYD
 
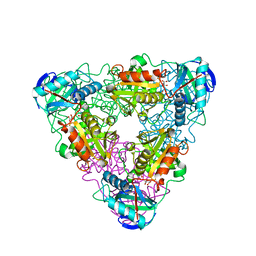 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, GLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hirotsu, K, Goto, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of branched-chain amino Acid aminotransferase complexed with glutamate and glutarate: true reaction intermediate and double substrate recognition of the enzyme.
Biochemistry, 42, 2003
|
|
5AVO
 
 | |
1X28
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1X29
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-2-methyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-2-METHYL-L-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
4YDR
 
 | |
8HR4
 
 | |
5X9D
 
 | | Crystal structure of homoserine dehydrogenase in complex with L-cysteine and NAD | | Descriptor: | (2R)-3-[[(4S)-3-aminocarbonyl-1-[(2R,3R,4S,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4H-pyridin-4-yl]sulfanyl]-2-azanyl-propanoic acid, Homoserine dehydrogenase, L(+)-TARTARIC ACID | | Authors: | Goto, M, Ogata, K, Kaneko, R, Yoshimune, K. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of homoserine dehydrogenase by formation of a cysteine-NAD covalent complex
Sci Rep, 8, 2018
|
|
1X2A
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-D-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
2YXZ
 
 | |
2DKJ
 
 | | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, serine hydroxymethyltransferase | | Authors: | Kai, K, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase
To be Published
|
|
3AB8
 
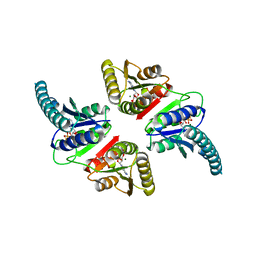 | |
2ZPU
 
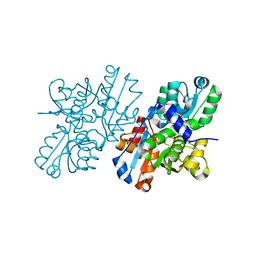 | | Crystal Structure of Modified Serine Racemase from S.pombe. | | Descriptor: | MAGNESIUM ION, N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, Uncharacterized protein C320.14 | | Authors: | Goto, M. | | Deposit date: | 2008-07-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serine racemase with catalytically active lysinoalanyl residue.
J.Biochem., 145, 2009
|
|
3AB7
 
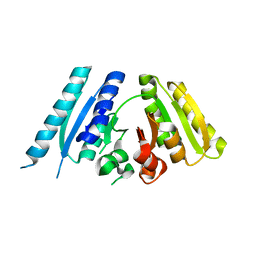 | |
1IQ0
 
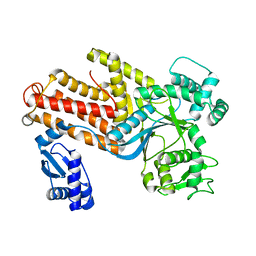 | | THERMUS THERMOPHILUS ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGINYL-TRNA SYNTHETASE | | Authors: | Shimada, A, Nureki, O, Goto, M, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7YQA
 
 | | Crystal structure of D-threonine aldolase from Chlamydomonas reinhardtii | | Descriptor: | D-threonine aldolase, MAGNESIUM ION | | Authors: | Hirato, Y, Goto, M, Mizobuchi, T, Muramatsu, H, Tanigawa, M, Nishimura, K. | | Deposit date: | 2022-08-05 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of pyridoxal 5'-phosphate-bound D-threonine aldolase from Chlamydomonas reinhardtii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
1WUO
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81A) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
7COK
 
 | | Crystal structure of ligand-free form of 5-ketofructose reductase of Gluconobacter sp. strain CHM43 | | Descriptor: | 5-ketofructose reductase | | Authors: | Noda, S, Hodoya, Y, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
7COL
 
 | | Crystal structure of 5-ketofructose reductase complexed with NADPH | | Descriptor: | 5-ketofructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hodoya, Y, Noda, S, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
6IUE
 
 | | DNA helical wire containing Hg(II) | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Ono, A, Kanazawa, H, Ito, H, Goto, M, Nakamura, K, Saneyoshi, H, Kondo, J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7CT4
 
 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7F4B
 
 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F4C
 
 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
