8XD7
 
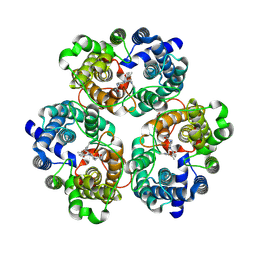 | |
8XDC
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDI
 
 | | Cryo-EM structure of zebrafish urea transporter. | | Descriptor: | N-(4-acetamidophenyl)-5-ethanoyl-furan-2-carboxamide, Urea transporter | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
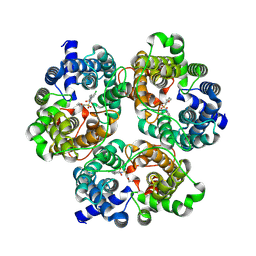
8XDB
 
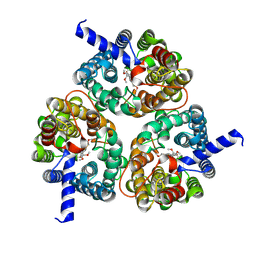 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | (5E)-2-azanylidene-5-[(2,3-dimethoxyphenyl)methylidene]-1,3-thiazolidin-4-one, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
5CC2
 
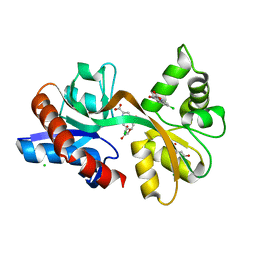 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
6Q54
 
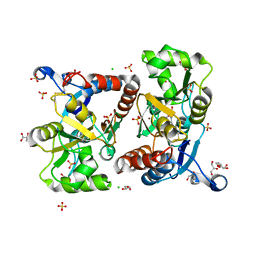 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(1-ethyl-4-hydroxy-1H-1,2,3-triazol-5-yl)propanoic acid at 1.4 A resolution | | Descriptor: | (2~{S})-2-azanyl-3-(3-ethyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Moellerud, S, Temperini, P, Kastrup, J.S. | | Deposit date: | 2018-12-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
6Q60
 
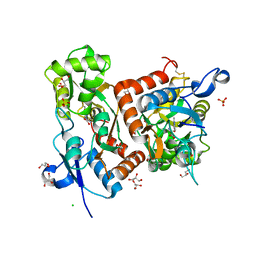 | | Structure of GluA2 ligand-binding domain (S1S2J) in complex with the agonist (S)-2-Amino-3-(2-methyl-5-hydroxy-2H-1,2,3-triazol-4-yl)propanoic acid at 1.55 A resolution | | Descriptor: | (2~{S})-2-azanyl-3-(2-methyl-5-oxidanyl-1,2,3-triazol-4-yl)propanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Moellerud, S, Temperini, P, Kastrup, J.S. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Use of the 4-Hydroxytriazole Moiety as a Bioisosteric Tool in the Development of Ionotropic Glutamate Receptor Ligands.
J.Med.Chem., 62, 2019
|
|
8DY2
 
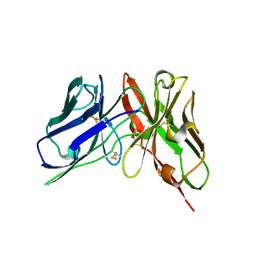 | | Crystal Structure of spFv GLK1 | | Descriptor: | SULFATE ION, spFv GLK1 LH | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY4
 
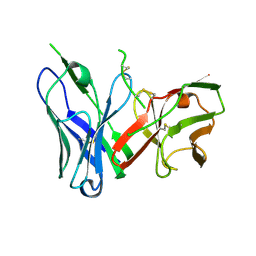 | | Crystal Structure of spFv CAT2200 HL | | Descriptor: | spFv CAT2200 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY5
 
 | |
8DY0
 
 | | Crystal Structure of spFv GLK1 HL | | Descriptor: | spFv GLK1 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY3
 
 | | Crystal Structure of spFv GLK2 HL | | Descriptor: | SULFATE ION, spFv GLK2 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY1
 
 | |
9C3E
 
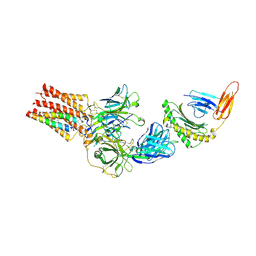 | | TCR - CD3 complex bound to HLA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Cancer/testis antigen 1,Beta-2-microglobulin,MHC class I antigen, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-05-31 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
8TW4
 
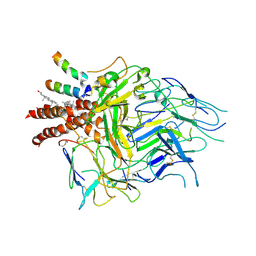 | | TCR in nanodisc ND-I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
8TW6
 
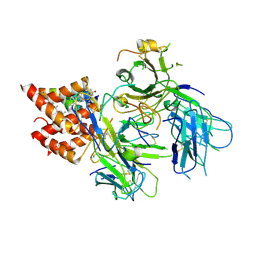 | | TCR in nanodisc ND-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, T cell receptor beta variable 6-5,T cell receptor beta chain MC.7.G5,MCHERRY, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2023-08-20 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
9BBC
 
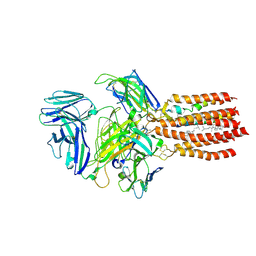 | | TCR GDN detergent micelle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Notti, R.Q, Walz, T. | | Deposit date: | 2024-04-05 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex.
Biorxiv, 2024
|
|
7XKE
 
 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
