8XBD
 
 | |
8XHO
 
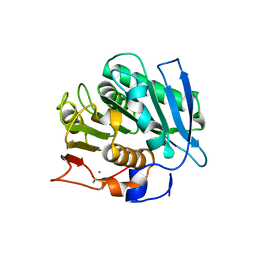 | |
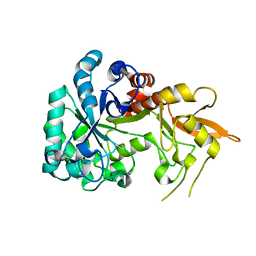
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8H5T
 
 | |
7SXO
 
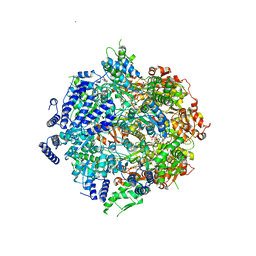 | | Yeast Lon (PIM1) with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-12 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
2LR7
 
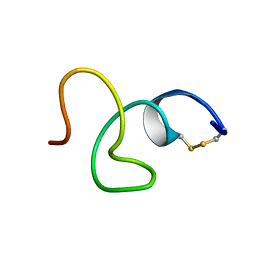 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
8J0P
 
 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JH5
 
 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JIA
 
 | | Marine bacterial laminarinase mutant E135A complex with laminaritetraose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
8JDI
 
 | |
8JDH
 
 | |
5YOX
 
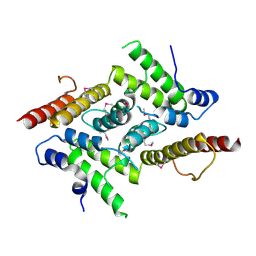 | | HD domain-containing protein YGK1(YGL101W) | | Descriptor: | HD domain-containing protein YGL101W, ZINC ION | | Authors: | Yang, J, Wang, F, Gao, Z, Zhou, K, Liu, Q. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | HD domain-containing protein YGK1(YGL101W)
To Be Published
|
|
7VW2
 
 | |
7VW1
 
 | |
7VW0
 
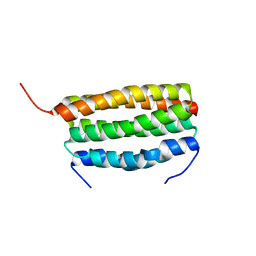 | | Structure of a dimeric periplasmic protein | | Descriptor: | DUF305 domain-containing protein | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle.
J.Inorg.Biochem., 229, 2022
|
|
8IOM
 
 | |
7VQM
 
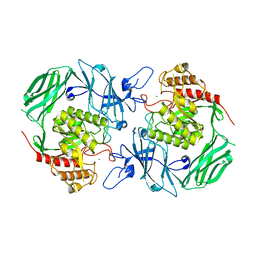 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
7CCB
 
 | |
7ECC
 
 | | M4 family peptidase PlM4P-mature form | | Descriptor: | CALCIUM ION, M4 family peptidase, PHOSPHATE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | M4 family peptidase PlM4P-mature form
To Be Published
|
|
7D6M
 
 | |
7D6N
 
 | |
7F0Y
 
 | |
7F13
 
 | | Crystal structure of isomerase Dcr3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dcr3 | | Authors: | Yang, J, Mori, T, Abe, I. | | Deposit date: | 2021-06-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Isomerization Reactions in Fungal Tetrahydroxanthone Biosynthesis and Diversification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
