5T1K
 
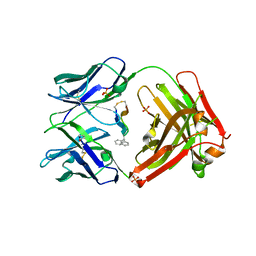 | | Cetuximab Fab in complex with CQFDA(Ph)2STRRLKC | | Descriptor: | CETUXIMAB FAB HEAVY CHAIN, CETUXIMAB FAB LIGHT CHAIN, CQFDA(PH)2STRRLKC PEPTIDE, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5T1L
 
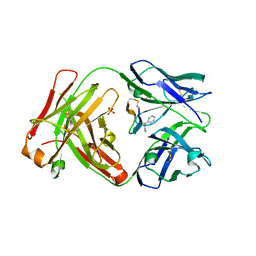 | | Cetuximab Fab in complex with CQA(Ph)2DLSTRRLKC peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CETUXIMAB FAB HEAVY CHAIN, CETUXIMAB FAB LIGHT CHAIN, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ICX
 
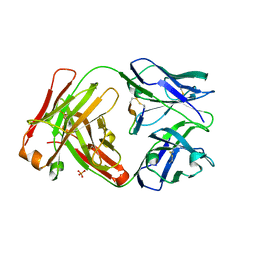 | | Cetuximab Fab in complex with CQFDLSTRRLRCGGSK meditope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ID1
 
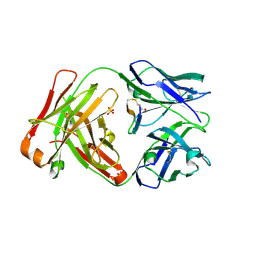 | | Cetuximab Fab in complex with MPT-Cys meditope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ICY
 
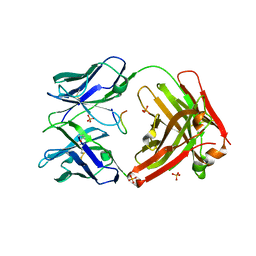 | | Cetuximab Fab in complex with linear meditope | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, Meditope, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5IOP
 
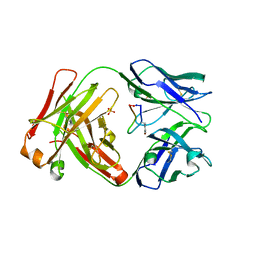 | |
5ICZ
 
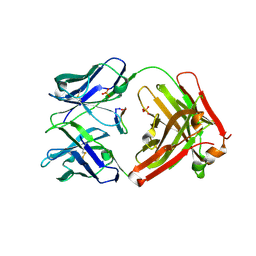 | | Cetuximab Fab in complex with GQFDLSTRRLKG peptide | | Descriptor: | Cetuximab Fab heavy chain, Cetuximab Fab light chain, Meditope, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5J9C
 
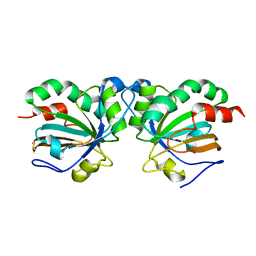 | | Crystal structure of peroxiredoxin Asp f3 C31S/C61S variant | | Descriptor: | MAGNESIUM ION, peroxiredoxin Asp f3 | | Authors: | Bzymek, K.P, Williams, J.C, Hong, T.B, Bagramyan, K, Kalkum, M. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | The Crystal Structure of Peroxiredoxin Asp f3 Provides Mechanistic Insight into Oxidative Stress Resistance and Virulence of Aspergillus fumigatus.
Sci Rep, 6, 2016
|
|
5IV2
 
 | | Cetuximab Fab in complex with Arg9Cir meditope variant | | Descriptor: | Cetuximab Fab, heavy chain, light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5J9B
 
 | | Crystal structure of peroxiredoxin Asp f3 | | Descriptor: | peroxiredoxin Asp f3 | | Authors: | Bzymek, K.P, Williams, J.C, Hong, T.B, Bagramyan, K, Kalkum, M. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Peroxiredoxin Asp f3 Provides Mechanistic Insight into Oxidative Stress Resistance and Virulence of Aspergillus fumigatus.
Sci Rep, 6, 2016
|
|
5ID0
 
 | | Cetuximab Fab in complex with aminoheptanoic acid-linked meditope | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Cyclization strategies of meditopes: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5ITF
 
 | |
5IVZ
 
 | | Cetuximab Fab in complex with Arg8Cir meditope variant | | Descriptor: | Cetuximab Fab, heavy chain, light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-03-21 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5IR1
 
 | | Cetuximab Fab in complex with 3-bromophenylalanine meditope variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab heavy chain, Cetuximab Fab light chain, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-03-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7RIK
 
 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
7JKM
 
 | |
1M3X
 
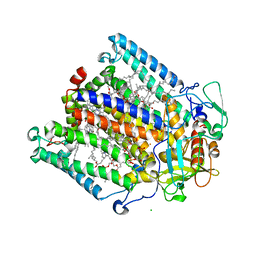 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Camara-Artigas, A, Brune, D, Allen, J.P. | | Deposit date: | 2002-07-01 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Interactions between lipids and bacterial reaction centers determined by protein crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1EAT
 
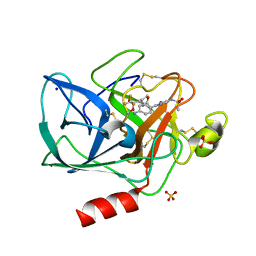 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 5. DESIGN, SYNTHESIS, AND X-RAY CRYSTALLOGRAPHY OF A SERIES OF ORALLY ACTIVE 5-AMINO-PYRIMIDIN-6-ONE-CONTAINING TRIFLUOROMETHYLKETONES | | Descriptor: | 2-[5-METHANESULFONYLAMINO-2-(4-AMINOPHENYL)-6-OXO-1,6-DIHYDRO-1-PYRIMIDINYL]-N-(3,3,3-TRIFLUORO-1-ISOPROPYL-2-OXOPROPYL)ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 5. Design, synthesis, and X-ray crystallography of a series of orally active 5-aminopyrimidin-6-one-containing trifluoromethyl ketones.
J.Med.Chem., 38, 1995
|
|
1EAU
 
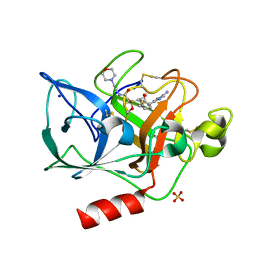 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 6. DESIGN OF A POTENT, INTRATRACHEALLY ACTIVE, PYRIDONE-BASED TRIFLUOROMETHYL KETONE | | Descriptor: | 2-[5-AMINO-6-OXO-2-(2-THIENYL)-1,6-DIHYDROPYRIMIDIN-1-YL)-N-[3,3-DIFLUORO -1-ISOPROPYL-2-OXO-3-(N-(2-MORPHOLINO ETHYL)CARBAMOYL]PROPYL]ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 6. Design of a potent, intratracheally active, pyridone-based trifluoromethyl ketone.
J.Med.Chem., 38, 1995
|
|
1EAS
 
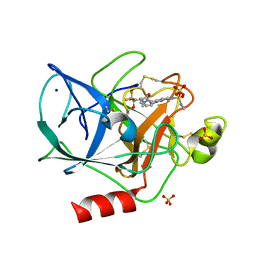 | | NONPEPTIDIC INHIBITORS OF HUMAN LEUKOCYTE ELASTASE. 3. DESIGN, SYNTHESIS, X-RAY CRYSTALLOGRAPHIC ANALYSIS, AND STRUCTURE-ACTIVITY RELATIONSHIPS FOR A SERIES OF ORALLY ACTIVE 3-AMINO-6-PHENYLPYRIDIN-2-ONE TRIFLUOROMETHYL KETONES | | Descriptor: | 3-[[(METHYLAMINO)SULFONYL]AMINO]-2-OXO-6-PHENYL-N-[3,3,3-TRIFLUORO-1-(1-METHYLETHYL)-2-OXOPHENYL]-1(2H)-PYRIDINE ACETAMIDE, PORCINE PANCREATIC ELASTASE, SODIUM ION, ... | | Authors: | Ceccarelli, C. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonpeptidic inhibitors of human leukocyte elastase. 3. Design, synthesis, X-ray crystallographic analysis, and structure-activity relationships for a series of orally active 3-amino-6-phenylpyridin-2-one trifluoromethyl ketones.
J.Med.Chem., 37, 1994
|
|
2MPX
 
 | |
1Z9K
 
 | |
7UZ9
 
 | |
7UZ8
 
 | |
7UZA
 
 | |
