3CFA
 
 | |
3W28
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W29
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W27
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W25
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3E5T
 
 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3W26
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W24
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
2Y0F
 
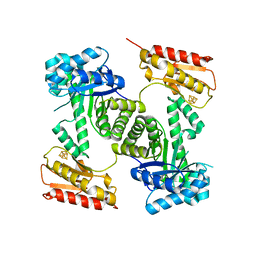 | | STRUCTURE OF GCPE (IspG) FROM THERMUS THERMOPHILUS HB27 | | Descriptor: | 4-HYDROXY-3-METHYLBUT-2-EN-1-YL DIPHOSPHATE SYNTHASE, IRON/SULFUR CLUSTER | | Authors: | Rekittke, I, Nonaka, T, Wiesner, J, Demmer, U, Warkentin, E, Jomaa, H, Ermler, U. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the E-1-Hydroxy-2-Methyl-But-2-Enyl-4-Diphosphate Synthase (Gcpe) from Thermus Thermophilus.
FEBS Lett., 585, 2011
|
|
2VVJ
 
 | | IrisFP fluorescent protein in its red form, cis conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3E5V
 
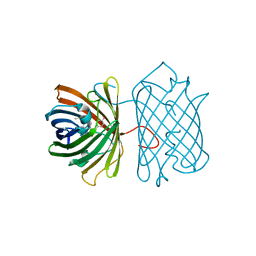 | | Crystal Structure Analysis of eqFP611 Double Mutant T122R, N143S | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
1ZUX
 
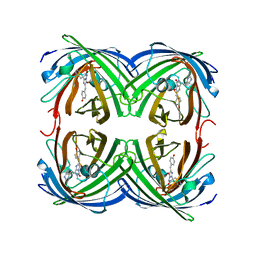 | | EosFP Fluorescent Protein- Green Form | | Descriptor: | green to red photoconvertible GPF-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C9J
 
 | | Structure of the fluorescent protein cmFP512 at 1.35A from Cerianthus membranaceus | | Descriptor: | GREEN FLUORESCENT PROTEIN FP512 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2006-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploring Chromophore-Protein Interactions in Fluorescent Protein Cmfp512 from Cerianthus Membranaceus: X-Ray Structure Analysis and Optical Spectroscopy.
Biochemistry, 45, 2006
|
|
2C9I
 
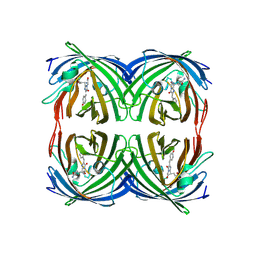 | | Structure of the fluorescent protein asFP499 from Anemonia sulcata | | Descriptor: | GREEN FLUORESCENT PROTEIN ASFP499 | | Authors: | Renzi, F, Nienhaus, K, Wiedenmann, J, Vallone, B, Nienhaus, G.U. | | Deposit date: | 2005-12-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chromophore-Protein Interactions in the Anthozoan Green Fluorescent Protein Asfp499
Biophys.J., 91, 2006
|
|
3VST
 
 | | The complex structure of XylC with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Xylosidase | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VSV
 
 | | The complex structure of XylC with xylose | | Descriptor: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VSU
 
 | | The complex structure of XylC with xylobiose | | Descriptor: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
1T1R
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, SULFATE ION, [(ISOQUINOLIN-1-YLAMINO)-PHOSPHONO-METHYL]-PHOSPHONIC ACID | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
8OR8
 
 | |
3WG9
 
 | | Crystal structure of RSP, a Rex-family repressor | | Descriptor: | Redox-sensing transcriptional repressor rex, SULFATE ION | | Authors: | Zheng, Y, Ko, T.-P, Guo, R.-T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Distinct structural features of Rex-family repressors to sense redox levels in anaerobes and aerobes.
J.Struct.Biol., 188, 2014
|
|
4IQB
 
 | | High Resolution Crystal Structure of C.elegans Thymidylate Synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
5BY6
 
 | | Crystal structure of Trichinella spiralis thymidylate synthase complexed with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Fraczyk, T, Wilk, P, Rode, W. | | Deposit date: | 2015-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
5NOO
 
 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP and Tomudex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Wilk, P, Jarmula, A, Maj, P, Dowiercial, A, Banaszak, K, Rypniewski, W, Rode, W. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
