4AAA
 
 | | Crystal structure of the human CDKL2 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CYCLIN-DEPENDENT KINASE-LIKE 2 | | Authors: | Canning, P, Vollmar, M, Cooper, C.D.O, Mahajan, P, Daga, N, Berridge, G, Burgess-Brown, N, Muniz, J.R.C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
3ZDU
 
 | | Crystal structure of the human CDKL3 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-DEPENDENT KINASE-LIKE 3, SODIUM ION, ... | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Pike, A.C.W, Quigley, A, MacKenzie, A, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
6NDZ
 
 | | Designed repeat protein in complex with Fz8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ACETIC ACID, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NE2
 
 | | Designed repeat protein in complex with Fz7 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Miao, Y, Jude, K.M, Garcia, K.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Receptor subtype discrimination using extensive shape complementary designed interfaces.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4BBM
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CDKL2 KINASE DOMAIN WITH BOUND TCS 2312 | | Descriptor: | 1,2-ETHANEDIOL, 4'-[5-[[3-[(CYCLOPROPYLAMINO)METHYL]PHENYL]AMINO]-1H-PYRAZOL-3-YL]-[1,1'-BIPHENYL]-2,4-DIOL, CHLORIDE ION, ... | | Authors: | Canning, P, Elkins, J.M, Cooper, C.D.O, Mahajan, P, Daga, N, Berridge, G, Burgess-Brown, N, Muniz, J.R.C, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A. | | Deposit date: | 2012-09-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
4BGQ
 
 | | Crystal structure of the human CDKL5 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYCLIN-DEPENDENT KINASE-LIKE 5, ... | | Authors: | Canning, P, Krojer, T, Goubin, S, Mahajan, P, Vollmar, M, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
3CI1
 
 | | Structure of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with four-coordinate cob(II)alamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | Maurice, M.St, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate.
Biochemistry, 47, 2008
|
|
3CI4
 
 | | Structure of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with four-coordinate cob(II)inamide and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COB(II)INAMIDE, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | Maurice, M.St, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate.
Biochemistry, 47, 2008
|
|
4KMF
 
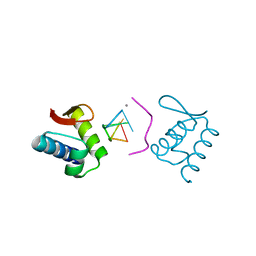 | |
5FS0
 
 | |
3RFJ
 
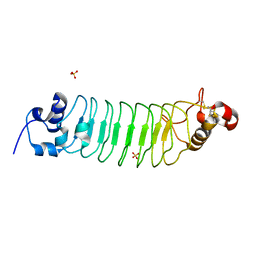 | | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering | | Descriptor: | Internalin B, repeat modules, Variable lymphocyte receptor, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-04-06 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RFS
 
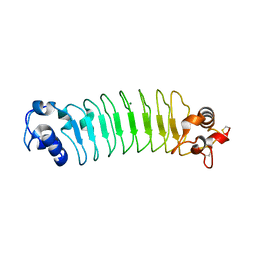 | | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering | | Descriptor: | Internalin B, repeat modules, Variable lymphocyte receptor B, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-04-06 | | Release date: | 2012-03-14 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UL8
 
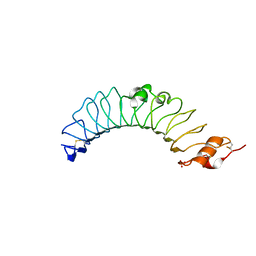 | | Crystal structure of the TV3 mutant V134L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
6L3T
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the absence of calcium | | Descriptor: | Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L3V
 
 | | The R15G mutant of human Cx31.3/GJC3 connexin hemichannel | | Descriptor: | Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L3U
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the presence of calcium | | Descriptor: | CALCIUM ION, Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6N4N
 
 | | Crystal structure of the designed protein DNCR2/danoprevir/NS3a complex | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, Rosetta-designed danoprevir/NS3a complex reader 2, ... | | Authors: | Wang, Z, Foight, G.W, Baker, D, Maly, D.J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Multi-input chemical control of protein dimerization for programming graded cellular responses.
Nat.Biotechnol., 37, 2019
|
|
6T96
 
 | | Photorhabdus laumondii subsp. laumondii lectin PLL3 | | Descriptor: | Lectin PLL3, SODIUM ION | | Authors: | Melicher, F, Houser, J, Fujdiarova, E, Faltinek, L, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lectin PLL3, a Novel Monomeric Member of the Seven-Bladed beta-Propeller Lectin Family.
Molecules, 24, 2019
|
|
6FHY
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{S},4~{R},6~{R})-2-(hydroxymethyl)-6-oxidanyl-oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6FHX
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{R},3~{S},4~{R},6~{R})-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6FLU
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{R},3~{S},4~{R},6~{S})-2-(hydroxymethyl)-6-methoxy-3-oxidanyl-oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Photorhabdus asymbitoca lectin PHL, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
1PPE
 
 | |
7FE1
 
 | | Crystal structure of GH92 alpha-1,2-mannosidase from Enterococcus faecalis ATCC 10100 in complex with methyl alpha-1,2-C-mannobioside | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Alpha-1,2-mannosidase, ... | | Authors: | Miyazaki, T, Alonso-Gil, S. | | Deposit date: | 2021-07-19 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Unlocking the Hydrolytic Mechanism of GH92 alpha-1,2-Mannosidases: Computation Inspires the use of C-Glycosides as Michaelis Complex Mimics.
Chemistry, 28, 2022
|
|
7FE2
 
 | | Crystal structure of the mutant E494Q of GH92 alpha-1,2-mannosidase from Enterococcus faecalis ATCC 10100 in complex with alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-1,2-mannosidase, ... | | Authors: | Miyazaki, T, Alonso-Gil, S. | | Deposit date: | 2021-07-19 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unlocking the Hydrolytic Mechanism of GH92 alpha-1,2-Mannosidases: Computation Inspires the use of C-Glycosides as Michaelis Complex Mimics.
Chemistry, 28, 2022
|
|
