7LIW
 
 | | Local refinement of human ATP citrate lyase E599Q mutant ASH domain | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Reply to: Acetyl-CoA is produced by the citrate synthase homology module of ATP-citrate lyase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5ICW
 
 | | Crystal structure of human NatF (hNaa60) homodimer bound to Coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, N-alpha-acetyltransferase 60 | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
1YGH
 
 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1ZX2
 
 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
6C95
 
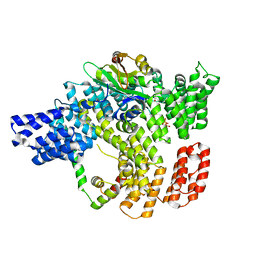 | |
6C9M
 
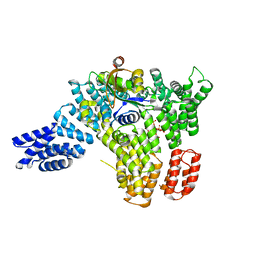 | |
6C9D
 
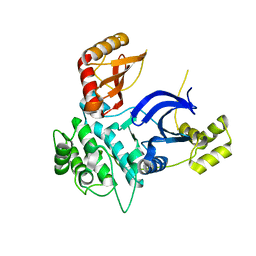 | |
1BC7
 
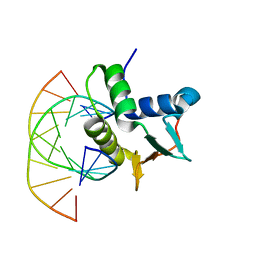 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1999-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
1BC8
 
 | | STRUCTURES OF SAP-1 BOUND TO DNA SEQUENCES FROM THE E74 AND C-FOS PROMOTERS PROVIDE INSIGHTS INTO HOW ETS PROTEINS DISCRIMINATE BETWEEN RELATED DNA TARGETS | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*TP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*AP*AP*GP*T)-3'), PROTEIN (SAP-1 ETS DOMAIN), ... | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1998-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
7UAG
 
 | | Structure of G6PD-WT dimer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of G6PD-WT dimer
To Be Published
|
|
7TOF
 
 | | Structure of G6PD-WT dimer with no symmetry applied | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TOE
 
 | |
7UC2
 
 | |
7UAL
 
 | |
1ZME
 
 | | CRYSTAL STRUCTURE OF PUT3/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*AP*GP*(5IU)P*TP*GP*GP*CP*TP*(5IU)P*CP*CP*CP*G)-3'), DNA (5'-D(*AP*CP*GP*GP*GP*AP*AP*GP*CP*CP*AP*AP*CP*TP*CP*CP*G)-3'), PROLINE UTILIZATION TRANSCRIPTION ACTIVATOR, ... | | Authors: | Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
6POF
 
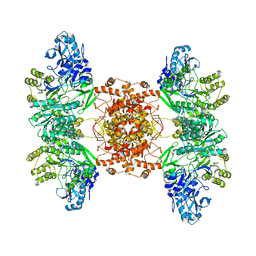 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PPL
 
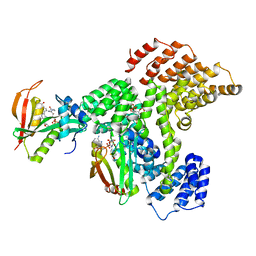 | | Cryo-EM structure of human NatE complex (NatA/Naa50) | | Descriptor: | ACETYL COENZYME *A, INOSITOL HEXAKISPHOSPHATE, N-alpha-acetyltransferase 10, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
6PW9
 
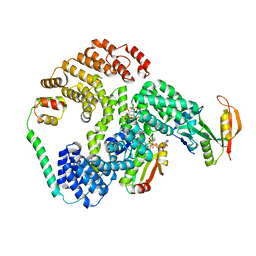 | | Cryo-EM structure of human NatE/HYPK complex | | Descriptor: | ACETYL COENZYME *A, Huntingtin-interacting protein K, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
4EQC
 
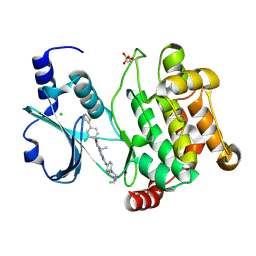 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
4GS4
 
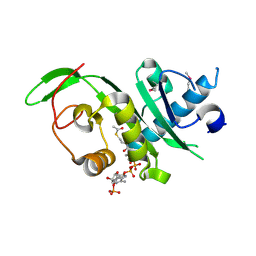 | | Structure of the alpha-tubulin acetyltransferase, alpha-TAT1 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase | | Authors: | Friedmann, D.R, Fan, J, Marmorstein, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Structure of the alpha-tubulin acetyltransferase, alpha TAT1, and implications for tubulin-specific acetylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4E26
 
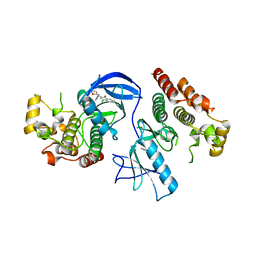 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
3TOB
 
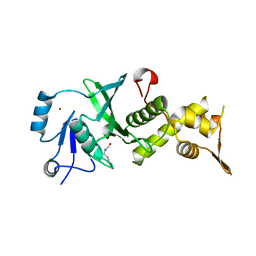 | |
3TO7
 
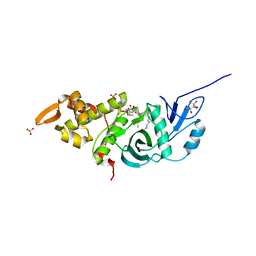 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
2B9D
 
 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
2ERE
 
 | |
