1SWP
 
 | | CORE-STREPTAVIDIN MUTANT W120F IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWR
 
 | | CORE-STREPTAVIDIN MUTANT W120A IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWL
 
 | | CORE-STREPTAVIDIN MUTANT W108F AT PH 7.0 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWG
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-07-12 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1SWF
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
3BWY
 
 | | Crystal Structure of Human 108M Catechol O-methyltransferase bound with S-adenosylmethionine and inhibitor dinitrocatechol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,5-DINITROCATECHOL, COMT protein, ... | | Authors: | Rutherford, K, Le Trong, I, Stenkamp, R.E, Parson, W.W. | | Deposit date: | 2008-01-10 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of human 108V and 108M catechol O-methyltransferase.
J.Mol.Biol., 380, 2008
|
|
3C5R
 
 | |
3I6A
 
 | | Human GST A1-1 GIMF mutant with Glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
3I69
 
 | | Apo Glutathione Transferase A1-1 GIMF-helix mutant | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Stenkamp, R.E, Atkins, W.M. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural analysis of a glutathione transferase A1-1 mutant tailored for high catalytic efficiency with toxic alkenals.
Biochemistry, 48, 2009
|
|
3IK9
 
 | | Human GST A1-1-GIMF with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
3IK7
 
 | | Human glutathione transferase a4-4 with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A4, SULFATE ION | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
2VER
 
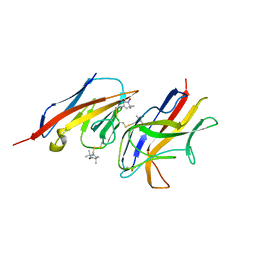 | | Structural model for the complex between the Dr adhesins and carcinoembryonic antigen (CEA) | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, ARCINOEMBRYONIC ANTIGEN-RELATED CELL ADHESION MOLECULE 5, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Korotkova, N, Yang, Y, Le Trong, I, Cota, E, Demeler, B, Marchant, J, Thomas, W.E, Stenkamp, R.E, Moseley, S.L, Matthews, S. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Binding of Dr Adhesins of Escherichia Coli to Carcinoembryonic Antigen Triggers Receptor Dissociation.
Mol.Microbiol., 67, 2008
|
|
3K34
 
 | | Human carbonic anhydrase II with a sulfonamide inhibitor | | Descriptor: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Behnke, C.A, Le Trong, I, Merritt, E.A, Teller, D.C, Stenkamp, R.E. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KPA
 
 | |
1EVU
 
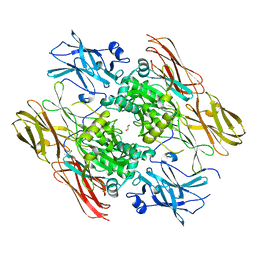 | | HUMAN FACTOR XIII WITH CALCIUM BOUND IN THE ION SITE | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII, S-1,2-PROPANEDIOL | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To Be Published
|
|
1EX0
 
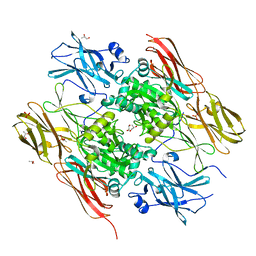 | | HUMAN FACTOR XIII, MUTANT W279F ZYMOGEN | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII A CHAIN, PHOSPHATE ION, ... | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-28 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To be Published
|
|
1QRK
 
 | | HUMAN FACTOR XIII WITH STRONTIUM BOUND IN THE ION SITE | | Descriptor: | PROTEIN (COAGULATION FACTOR XIII), STRONTIUM ION | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Le Trong, I, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
3RQ9
 
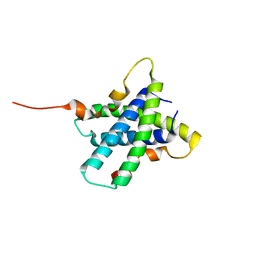 | | Structure of Tsi2, a Tse2-immunity protein from Pseudomonas aeruginosa | | Descriptor: | Type VI secretion immunity protein | | Authors: | Li, M, Le Trong, I, Stenkamp, R.E, Mougous, J.D. | | Deposit date: | 2011-04-27 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis for Type VI Secretion Effector Recognition by a Cognate Immunity Protein.
Plos Pathog., 8, 2012
|
|
3FA2
 
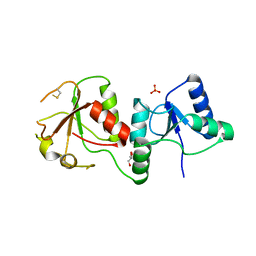 | |
3T6F
 
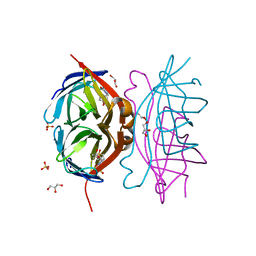 | | Biotin complex of Y54F core streptavidin | | Descriptor: | BIOTIN, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Baugh, L, Le Trong, I, Cerutti, D.S, Mehta, N, Gulich, S, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
3T6L
 
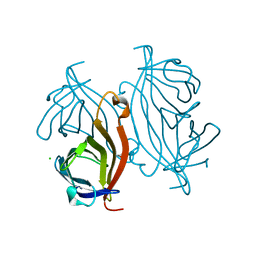 | | Y54F mutant of core streptavidin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Streptavidin | | Authors: | Baugh, L, Le Trong, I, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
1LUQ
 
 | |
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
