5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
6BX3
 
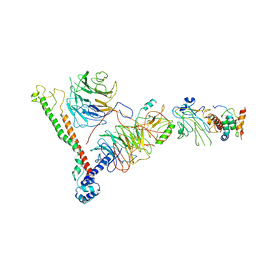 | | Structure of histone H3k4 methyltransferase | | Descriptor: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | Authors: | Skiniotis, G, Qu, Q.H. | | Deposit date: | 2017-12-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
5VA6
 
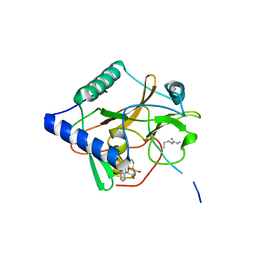 | | CRYSTAL STRUCTURE OF ATXR5 IN COMPLEX WITH HISTONE H3.1 MONO-METHYLATED ON R26 | | Descriptor: | Histone H3.1, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VAH
 
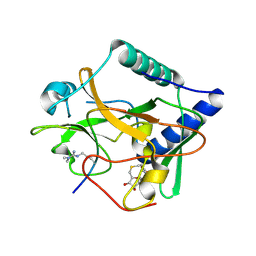 | | Crystal structure of ATXR5 SET domain in complex with histone H3 di-methylated on R26 | | Descriptor: | Histone H3.2, Probable Histone-lysine N-methyltransferase ATXR5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-26 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VBC
 
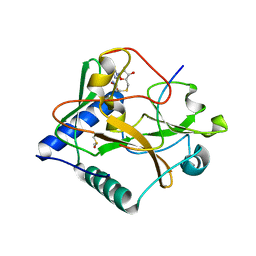 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5VAB
 
 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | Descriptor: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
3M53
 
 | | SET7/9 in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M55
 
 | | SET7/9 Y305F in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M58
 
 | | SET7/9 Y245A in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M5A
 
 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M56
 
 | | SET7/9 Y305F in complex with TAF10-K189me2 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10-K189me2 PEPTIDE | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M54
 
 | | SET7/9 Y305F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M57
 
 | | SET7/9 Y245A in complex with TAF10 peptide and AdoHcy | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M59
 
 | | SET7/9 Y245A in complex with TAF10-K189me2 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3SMR
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
8CUB
 
 | |
3IVS
 
 | | Homocitrate Synthase Lys4 | | Descriptor: | COBALT (II) ION, Homocitrate synthase, mitochondrial, ... | | Authors: | Bulfer, S.L, Scott, E.M, Couture, J.-F, Pillus, L, Trievel, R.C. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
J.Biol.Chem., 284, 2009
|
|
3IVU
 
 | | Homocitrate Synthase Lys4 bound to 2-OG | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, Homocitrate synthase, ... | | Authors: | Bulfer, S.L, Scott, E.M, Couture, J.-F, Pillus, L, Trievel, R.C. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
J.Biol.Chem., 284, 2009
|
|
3IVT
 
 | | Homocitrate Synthase Lys4 bound to 2-OG | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, mitochondrial, ... | | Authors: | Bulfer, S.L, Scott, E.M, Couture, J.-F, Pillus, L, Trievel, R.C. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
J.Biol.Chem., 284, 2009
|
|
