3MQT
 
 | |
3M0F
 
 | |
3M7V
 
 | | Crystal structure of phosphopentomutase from streptococcus mutans | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Fedorov, A.A, Bonanno, J, Fedorov, E.V, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopentomutase from streptococcus mutans
To be Published
|
|
3M8N
 
 | |
3MDK
 
 | |
3MC1
 
 | | Crystal structure of a predicted phosphatase from Clostridium acetobutylicum | | Descriptor: | CHLORIDE ION, GLYCEROL, Predicted phosphatase, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-26 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a predicted phosphatase from Clostridium acetobutylicum
To be Published, 2010
|
|
3M2P
 
 | |
3MF4
 
 | | Crystal structure of putative two-component system response regulator/ggdef domain protein | | Descriptor: | MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative two-component system response regulator/ggdef domain protein
To be Published
|
|
1NLW
 
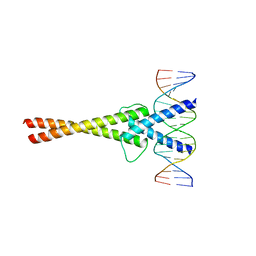 | | Crystal structure of Mad-Max recognizing DNA | | Descriptor: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1EJ1
 
 | | COCRYSTAL STRUCTURE OF THE MESSENGER RNA 5' CAP-BINDING PROTEIN (EIF4E) BOUND TO 7-METHYL-GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP.
Cell(Cambridge,Mass.), 89, 1997
|
|
1EJH
 
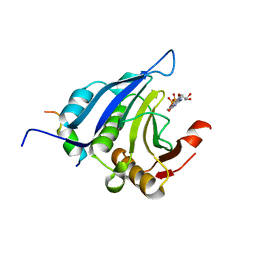 | | EIF4E/EIF4G PEPTIDE/7-METHYL-GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC INITIATION FACTOR 4GII | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-03-02 | | Release date: | 2000-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1G7S
 
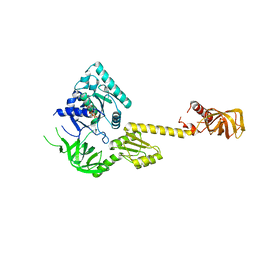 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1G61
 
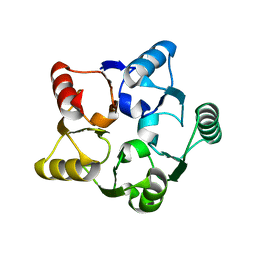 | | CRYSTAL STRUCTURE OF M.JANNASCHII EIF6 | | Descriptor: | TRANSLATION INITIATION FACTOR 6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1G7R
 
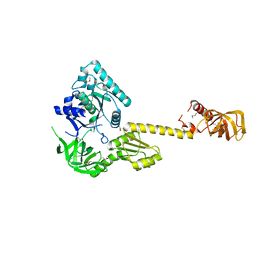 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B | | Descriptor: | TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1FI4
 
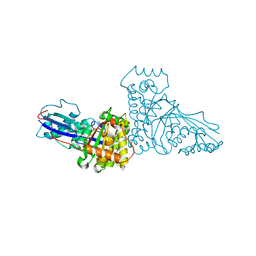 | | THE X-RAY CRYSTAL STRUCTURE OF MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AT 2.3 ANGSTROM RESOLUTION. | | Descriptor: | MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-08-03 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HU3
 
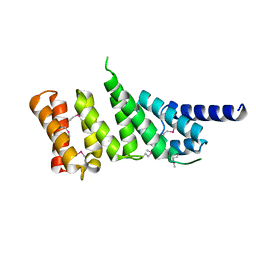 | | MIDDLE DOMAIN OF HUMAN EIF4GII | | Descriptor: | EIF4GII | | Authors: | Marcotrigiano, J, Lomakin, I, Pestova, T.V, Hellen, C.U.T, Sonenberg, N, Burley, S.K. | | Deposit date: | 2001-01-03 | | Release date: | 2001-02-21 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery.
Mol.Cell, 7, 2001
|
|
1G7T
 
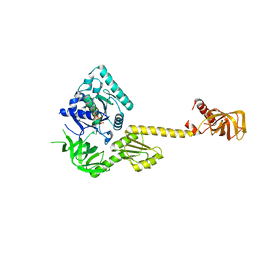 | | X-RAY STRUCTURE OF TRANSLATION INITIATION FACTOR IF2/EIF5B COMPLEXED WITH GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSLATION INITIATION FACTOR IF2/EIF5B | | Authors: | Roll-Mecak, A, Cao, C, Dever, T.E, Burley, S.K. | | Deposit date: | 2000-11-14 | | Release date: | 2000-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding.
Cell(Cambridge,Mass.), 103, 2000
|
|
1I9A
 
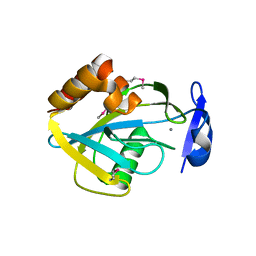 | | STRUCTURAL STUDIES OF CHOLESTEROL BIOSYNTHESIS: MEVALONATE 5-DIPHOSPHATE DECARBOXYLASE AND ISOPENTENYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Edo, C, Eswar, N, Pieper, U, Romanowski, M.J, Ilyin, V, Gerchman, S.E, Kycia, H, Studier, F.W, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-03-18 | | Release date: | 2001-03-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I2T
 
 | |
1G62
 
 | | CRYSTAL STRUCTURE OF S.CEREVISIAE EIF6 | | Descriptor: | RIBOSOME ANTI-ASSOCIATION FACTOR EIF6 | | Authors: | Groft, C.M, Beckmann, R, Sali, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of ribosome anti-association factor IF6.
Nat.Struct.Biol., 7, 2000
|
|
1I27
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF THE RAP74 SUBUNIT OF HUMAN TRANSCRIPTION FACTOR IIF (TFIIF) | | Descriptor: | TRANSCRIPTION FACTOR IIF, ZINC ION | | Authors: | Kamada, K, De Angelis, J, Roeder, R.G, Burley, S.K. | | Deposit date: | 2001-02-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the C-terminal domain of the RAP74 subunit of human transcription factor IIF.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
2G59
 
 | |
2F4N
 
 | |
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
