6VY8
 
 | |
6VXY
 
 | | Triazole bridged SFTI1 inhibitor in complex with beta-trypsin | | Descriptor: | 1-methyl-1H-1,2,3-triazole, CALCIUM ION, Cationic trypsin, ... | | Authors: | White, A.M, King, G.J, Durek, T, Craik, D.J. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 2020
|
|
4TTO
 
 | |
7JGY
 
 | |
7K7X
 
 | |
7JN6
 
 | |
1QH2
 
 | |
7JGX
 
 | |
7JHF
 
 | |
7KPD
 
 | |
7JNN
 
 | |
5KWO
 
 | |
5KWX
 
 | |
5KWP
 
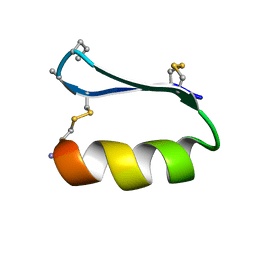 | |
5KVN
 
 | |
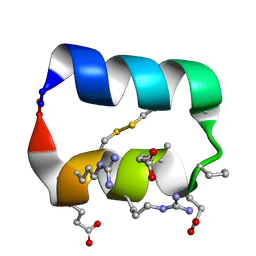
5KX0
 
 | |
5KX2
 
 | |
5KWZ
 
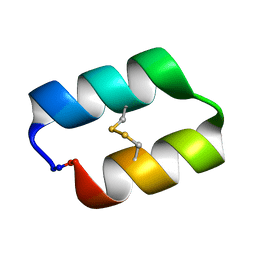 | |
5KX1
 
 | |
1WN8
 
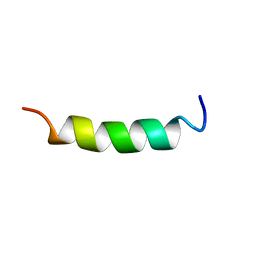 | | NMR Structure of OaNTR | | Descriptor: | Kalata B3/B6 | | Authors: | Dutton, J.L, Renda, R.F, Waine, C, Clark, R.J, Daly, N.L, Jennings, C.V, Anderson, M.A, Craik, D.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins
J.Biol.Chem., 279, 2004
|
|
1XGB
 
 | |
1XGC
 
 | |
1XGA
 
 | |
1YTP
 
 | |
7RMS
 
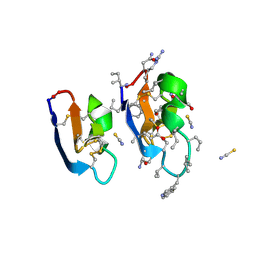 | | Crystal structure of [I11G]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
