5HZG
 
 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
4OBQ
 
 | | MAP4K4 in complex with inhibitor (compound 31), N-[3-(4-AMINOQUINAZOLIN-6-YL)-5-FLUOROPHENYL]-2-(PYRROLIDIN-1-YL)ACETAMIDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
7TBF
 
 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 antibody A19-61.1, Heavy chain of SARS-CoV-2 antibody B1-182.1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
7TB8
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
5HYW
 
 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
6O2Y
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 24 | | Descriptor: | 4-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methoxybenzonitrile, BETA-MERCAPTOETHANOL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
4ZSE
 
 | |
6XQR
 
 | | OXA-48 bound by Compound 2.2 | | Descriptor: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6P3P
 
 | | Crystal structure of Mcl-1 in complex with compound 65 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, methyl N-(5-{[2-chloro-5-(trifluoromethyl)phenyl]sulfamoyl}-4-methylthiophene-2-carbonyl)-D-phenylalaninate | | Authors: | Toms, A.V, Follows, B. | | Deposit date: | 2019-05-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of novel biaryl sulfonamide based Mcl-1 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3B9O
 
 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3B9N
 
 | | Crystal structure of long-chain alkane monooxygenase (LadA) | | Descriptor: | Alkane monooxygenase | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
5EA6
 
 | | Crystal Structure of Inhibitor BTA-9881 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | (9~{b}~{R})-9~{b}-(4-chlorophenyl)-1-pyridin-3-ylcarbonyl-2,3-dihydroimidazo[5,6]pyrrolo[1,2-~{a}]pyridin-5-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5EA5
 
 | | Crystal Structure of Inhibitor TMC-353121 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, D(-)-TARTARIC ACID, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
1CE9
 
 | | HELIX CAPPING IN THE GCN4 LEUCINE ZIPPER | | Descriptor: | PROTEIN (GCN4-PMSE) | | Authors: | Lu, M, Shu, W, Ji, H, Spek, E, Wang, L.-Y, Kallenbach, N.R. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Helix capping in the GCN4 leucine zipper.
J.Mol.Biol., 288, 1999
|
|
7SEL
 
 | | E. coli MsbA in complex with LPS and inhibitor G7090 (compound 3) | | Descriptor: | (2E)-3-{7-[(1S)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-1-methylnaphthalen-2-yl}prop-2-enoic acid, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Payandeh, J, Koth, C.M, Verma, V.A. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.978 Å) | | Cite: | Discovery of Inhibitors of the Lipopolysaccharide Transporter MsbA: From a Screening Hit to Potent Wild-Type Gram-Negative Activity.
J.Med.Chem., 65, 2022
|
|
3L8E
 
 | | Crystal Structure of apo form of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli | | Descriptor: | ACETIC ACID, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
4XAK
 
 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
3L8F
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with magnesium and phosphate | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3L8G
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with D-glycero-D-manno-heptose 1 ,7-bisphosphate | | Descriptor: | 1,7-di-O-phosphono-L-glycero-beta-D-manno-heptopyranose, D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3L8H
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from B. bronchiseptica complexed with magnesium and phosphate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
5H25
 
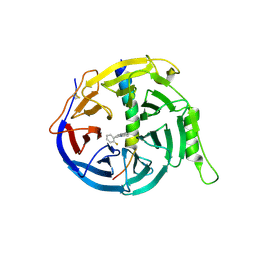 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
7VGG
 
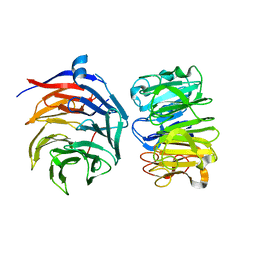 | | Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1 | | Descriptor: | E3 ubiquitin-protein ligase COP1, Ultraviolet-B receptor UVR8 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into UV-B-activated UVR8 bound to COP1.
Sci Adv, 8, 2022
|
|
5H24
 
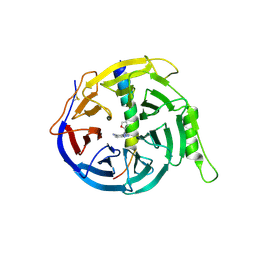 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6URM
 
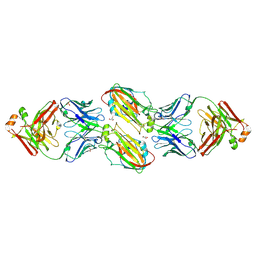 | | Crystal structure of vaccine-elicited receptor-binding site targeting antibody LPAF-a.01 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhou, T, Cheung, S.F, Kwong, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation.
Cell Rep, 32, 2020
|
|
