5HV1
 
 | |
6OWD
 
 | |
5HV6
 
 | |
5HV3
 
 | |
6OAZ
 
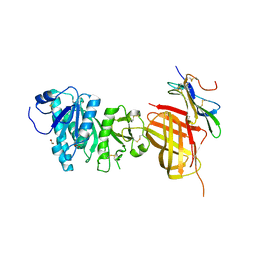 | | Apo Structure of WT Lipoprotein Lipase in Complex with GPIHBP1 Mutant N78D N82D produced in HEK293-F cells | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arora, R, Horton, P.A, Benson, T.E, Romanowski, M.J. | | Deposit date: | 2019-03-19 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of lipoprotein lipase in complex with GPIHBP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5Y9P
 
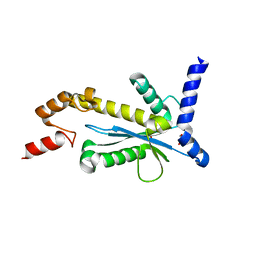 | | Staphylococcus aureus RNase HII | | Descriptor: | GLYCEROL, Ribonuclease HII | | Authors: | Hang, T, Wu, M, Zhang, X. | | Deposit date: | 2017-08-26 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a novel functional dimer of Staphylococcus aureus RNase HII
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5JD0
 
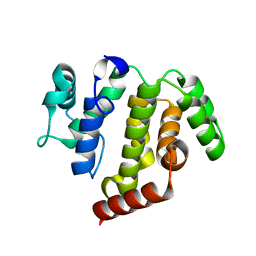 | | crystal structure of ARAP3 RhoGAP domain | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Bao, H, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
6KZ1
 
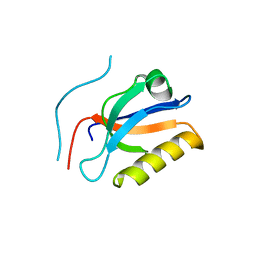 | | Complex structure of Whirlin and Myosin XVa | | Descriptor: | Myosin XVa, Whirlin | | Authors: | Lin, L, Wang, M, Shi, Y, Zhu, J, Zhang, R. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex.
Cell Rep, 34, 2021
|
|
6RTH
 
 | |
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
8FA2
 
 | |
8FA1
 
 | |
7DLV
 
 | | shrimp dUTPase in complex with Stl | | Descriptor: | CALCIUM ION, Orf20, SULFATE ION, ... | | Authors: | Ma, Q, Wang, F. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of staphylococcal Stl inhibition on a eukaryotic dUTPase.
Int.J.Biol.Macromol., 184, 2021
|
|
5T18
 
 | | Crystal structure of Bruton agammabulinemia tyrosine kinase complexed with BMS-986142 aka (2s)-6-fluoro-5-[3-(8-fluoro-1-methyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-3-yl)-2-methylphenyl]-2-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1h-carbazole-8-carboxamide | | Descriptor: | 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-08-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton's Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers.
J. Med. Chem., 59, 2016
|
|
8GDS
 
 | |
4KB5
 
 | | Crystal structure of MycP1 from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Tian, C.L. | | Deposit date: | 2013-04-23 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
6RTG
 
 | | Crystal structure of the UDP-bound glycosyltransferase domain from the YGT toxin | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Wirth, C, Bogdanovic, X, Kao, W.-C, Hunte, C. | | Deposit date: | 2019-05-23 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverse control of Rab proteins byYersiniaADP-ribosyltransferase and glycosyltransferase related to clostridial glucosylating toxins.
Sci Adv, 6, 2020
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7FD2
 
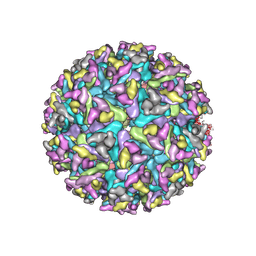 | | Cryo-EM structure of an alphavirus, Getah virus | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, Z, Liu, C, Wang, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of infective Getah virus at 2.8 angstrom resolution determined by cryo-electron microscopy.
Cell Discov, 8, 2022
|
|
6LEV
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 46 and NADPH | | Descriptor: | 2-[[4,6-bis(azanyl)-2,2-dimethyl-1,3,5-triazin-1-yl]oxy]-N-(4-chlorophenyl)ethanamide, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
7OLX
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
