4HC5
 
 | |
4JWO
 
 | | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776
To be Published
|
|
4GYQ
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase NDM-1, MAGNESIUM ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 D223A mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4KVF
 
 | | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836 | | Descriptor: | GLYCEROL, Rhamnose ABC transporter, periplasmic rhamnose-binding protein | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836
To be Published
|
|
4GQT
 
 | | N-terminal domain of C. elegans Hsp90 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90, ZINC ION | | Authors: | Osipiuk, J, Chhor, G, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-terminal domain of C. elegans Hsp90
To be Published
|
|
4GPN
 
 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4HYL
 
 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
3GX1
 
 | |
4HKU
 
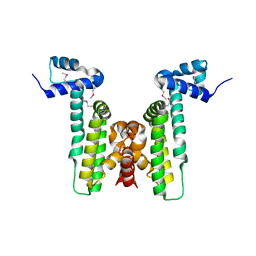 | |
4HTL
 
 | | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucoside kinase | | Authors: | Osipiuk, J, Mack, J, Endres, M, Salazar, J, Zhang, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Lmo2764 protein, a putative N-acetylmannosamine kinase, from Listeria monocytogenes.
To be Published
|
|
3HE1
 
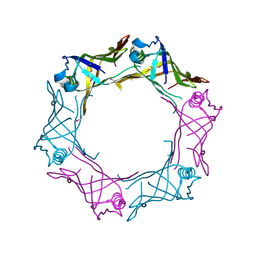 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
4EY3
 
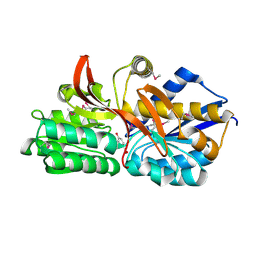 | | Crystal structure of solute binding protein of ABC transporter in complex with p-hydroxybenzoic acid | | Descriptor: | P-HYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
3HDJ
 
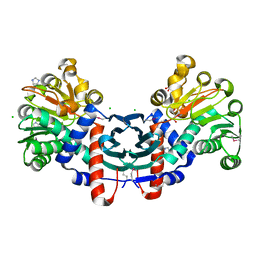 | | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I
To be Published
|
|
4Q2B
 
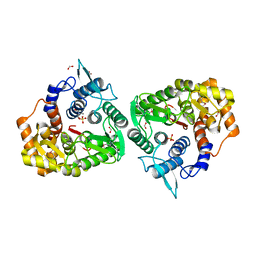 | | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-D-glucanase, FORMIC ACID, ... | | Authors: | Tan, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440
To be Published
|
|
3HIU
 
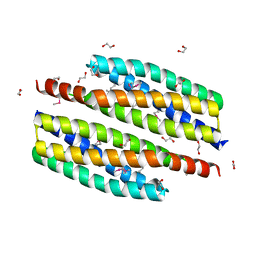 | | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of protein (XCC3681) from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
4Q52
 
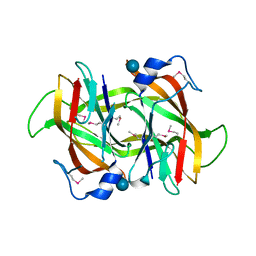 | | 2.60 Angstrom resolution crystal structure of a conserved uncharacterized protein from Chitinophaga pinensis DSM 2588 | | Descriptor: | Uncharacterized protein, beta-D-glucopyranose | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Minasov, G, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom resolution crystal structure of a conserved uncharacterized protein from Chitinophaga pinensis DSM 2588
To be Published
|
|
3HHL
 
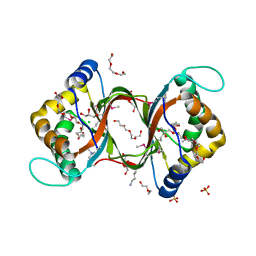 | | Crystal structure of methylated RPA0582 protein | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sledz, P, Niedzialkowska, E, Chruszcz, M, Porebski, P, Yim, V, Kudritska, M, Zimmerman, M.D, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of methylated RPA0582 protein
To be Published
|
|
4I6Z
 
 | | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide | | Descriptor: | DNA OLIGONUCLEOTIDE, Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide
To be Published
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4EYK
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with 3,4-dihydroxy benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4Q6B
 
 | | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular ligand-binding receptor, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu
To be Published, 2014
|
|
4EXK
 
 | | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 from Salmonella enterica | | Descriptor: | Maltose-binding periplasmic protein, uncharacterized protein chimera, TRIETHYLENE GLYCOL, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 form Salmonella enterica
To be Published
|
|
4R0J
 
 | | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein SMU1763c from Streptococcus mutans
To be Published
|
|
3LO3
 
 | |
