1HGX
 
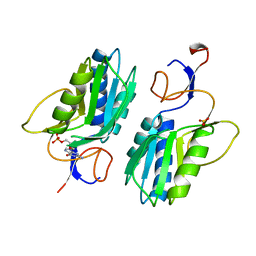 | |
6GNS
 
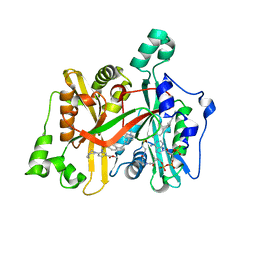 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[[4-[4-(1-methylpiperidin-4-yl)butyl]phenyl]sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
1HKY
 
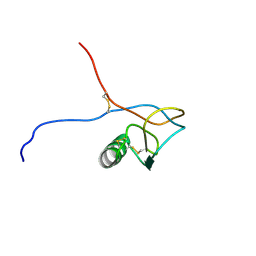 | | Solution structure of a PAN module from Eimeria tenella | | Descriptor: | MICRONEME PROTEIN 5 PRECURSOR | | Authors: | Brown, P.J, Mulvey, D, Potts, J.R, Tomley, F.M, Campbell, I.D. | | Deposit date: | 2002-10-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Pan Module from the Apicomplexan Parasite Eimeria Tenella
J.Struct.Funct.Genom., 4, 2003
|
|
1HRZ
 
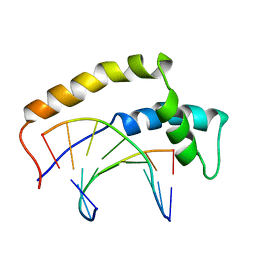 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
1C3T
 
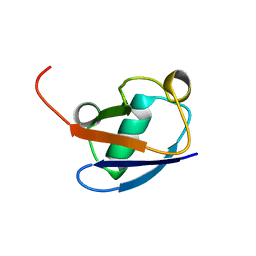 | |
6GV0
 
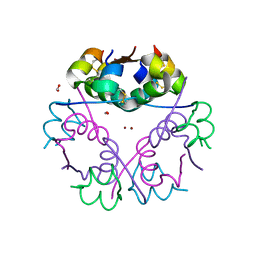 | | Insulin glulisine | | Descriptor: | FORMIC ACID, Insulin, ZINC ION | | Authors: | Chayen, N.E, Helliwell, J.R, Solomon-Gamsu, H.V, Govada, L, Morgan, M, Gillis, R.B, Adams, G. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Analysis of insulin glulisine at the molecular level by X-ray crystallography and biophysical techniques.
Sci Rep, 11, 2021
|
|
6GNU
 
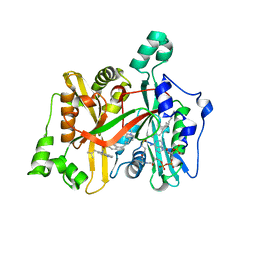 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-[6-(2-methylpropyl)quinolin-5-yl]benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GJU
 
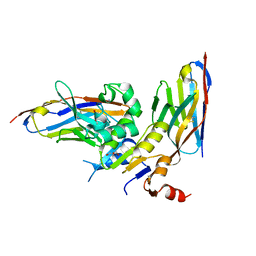 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6GNT
 
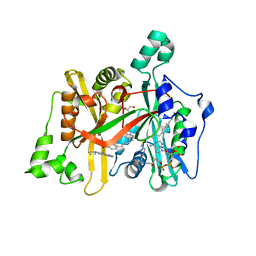 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a Quionlinyl aryl sulphonamide ligand | | Descriptor: | 4-[4-(1-methylpiperidin-4-yl)butyl]-~{N}-(6-pyrrolidin-1-ylquinolin-5-yl)benzenesulfonamide, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
1AJU
 
 | |
1QGB
 
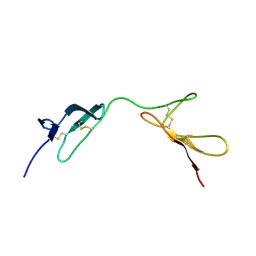 | | SOLUTION STRUCTURE OF THE N-TERMINAL F1 MODULE PAIR FROM HUMAN FIBRONECTIN | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Potts, J.R, Bright, J.R, Bolton, D, Pickford, A.R, Campbell, I.D. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal F1 module pair from human fibronectin.
Biochemistry, 38, 1999
|
|
1EHI
 
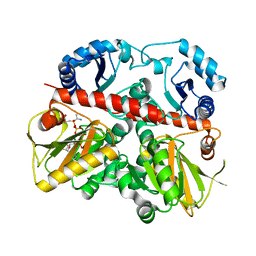 | | D-ALANINE:D-LACTATE LIGASE (LMDDL2) OF VANCOMYCIN-RESISTANT LEUCONOSTOC MESENTEROIDES | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE:D-LACTATE LIGASE, ... | | Authors: | Kuzin, A.P, Sun, T, Jorczak-Baillass, J, Healy, V.L, Walsh, C.T, Knox, J.R. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Enzymes of vancomycin resistance: the structure of D-alanine-D-lactate ligase of naturally resistant Leuconostoc mesenteroides.
Structure Fold.Des., 8, 2000
|
|
6OAA
 
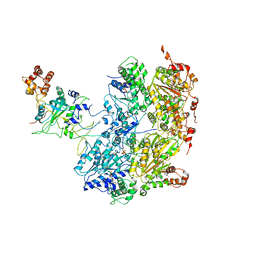 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OA9
 
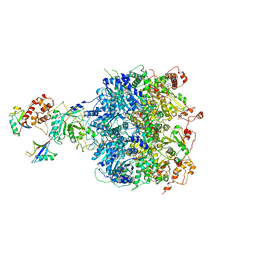 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K127L at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-04-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6P64
 
 | |
6PBY
 
 | |
6P7E
 
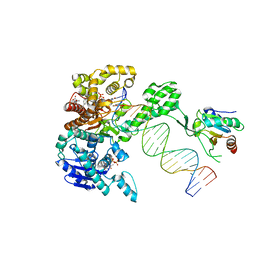 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
6PBX
 
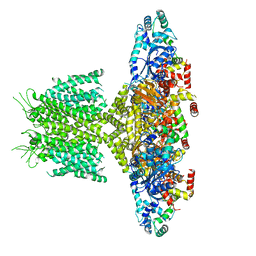 | |
6PBD
 
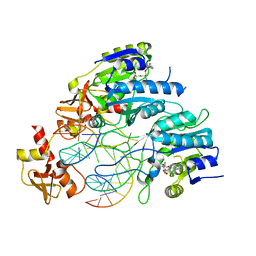 | | DNA N6-Adenine Methyltransferase CcrM In Complex with Double-Stranded DNA Oligonucleotide Containing Its Recognition Sequence GAATC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*AP*TP*TP*CP*AP*AP*TP*GP*AP*AP*TP*CP*CP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*GP*GP*AP*TP*TP*CP*AP*TP*TP*GP*AP*AP*TP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Woodcock, C.B. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site.
Nat Commun, 10, 2019
|
|
2HZ8
 
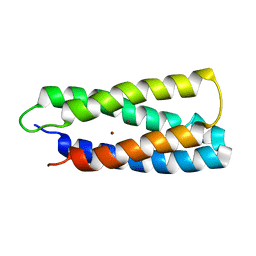 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
3ZF2
 
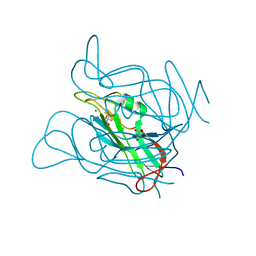 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
6OD5
 
 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
3ZF6
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A D110C S168C mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF1
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81N mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
