4LWH
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4RVZ
 
 | | Crystal structure of tRNA fluorescent labeling enzyme | | Descriptor: | MAGNESIUM ION, N-(4-aminobutyl)-2-azidoacetamide, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Dong, J, Li, F, Wang, J, Gong, W. | | Deposit date: | 2014-11-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A covalent approach for site-specific RNA labeling in Mammalian cells.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TV5
 
 | |
4UMX
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
4UMY
 
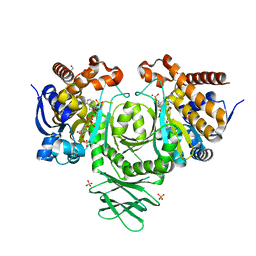 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McLean, L, Zhang, Y, Mathieu, M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
4TV6
 
 | |
5AA4
 
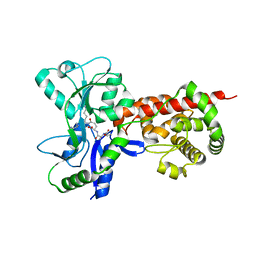 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide | | Descriptor: | MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, [6-[[(2~{R})-1-azanyl-1-oxidanylidene-propan-2-yl]amino]-6-oxidanylidene-5-[[(4~{R})-5-oxidanyl-5-oxidanylidene-4-[[(2~{S})-2-[[(2~{R})-2-oxidanylpropanoyl]amino]propanoyl]amino]pentanoyl]amino]hexyl]azanium | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
4QS9
 
 | |
4QS8
 
 | |
7U1Z
 
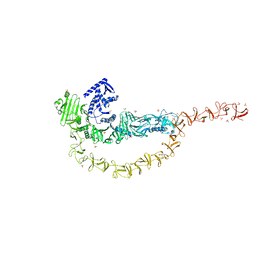 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7UBY
 
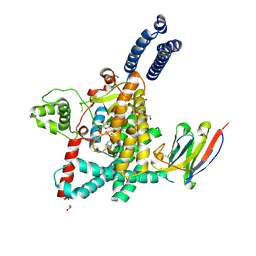 | | Structure of the GTD domain of Clostridium difficile toxin A in complex with VHH AH3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Glucosyltransferase TcdA, ... | | Authors: | Chen, B, Rongsheng, J, Kay, P. | | Deposit date: | 2022-03-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.
Front Immunol, 13, 2022
|
|
6CRP
 
 | |
6CSH
 
 | |
6CRR
 
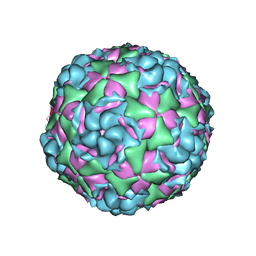 | |
6CSA
 
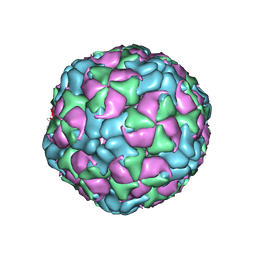 | |
6CSG
 
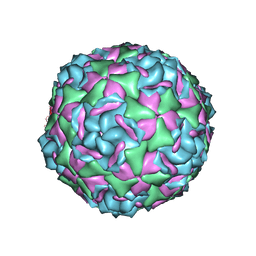 | |
6CS3
 
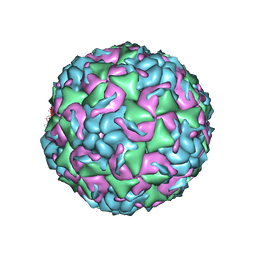 | |
6CS5
 
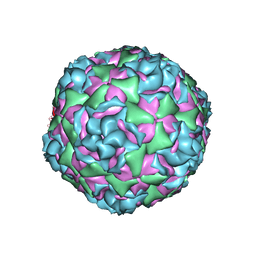 | |
8OJE
 
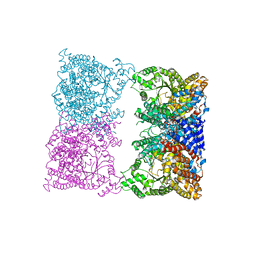 | |
6CRS
 
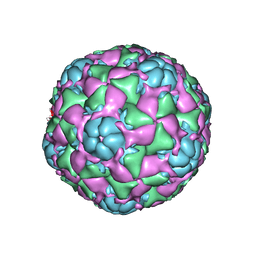 | |
8OJY
 
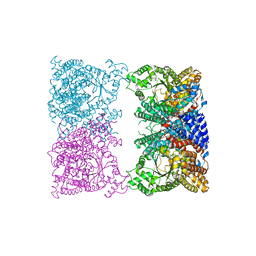 | |
6CRU
 
 | |
6CS4
 
 | |
6CS6
 
 | |
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
