7CVQ
 
 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
5LSY
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5LT6
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5XUU
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
3DK3
 
 | |
4YT3
 
 | | CYP106A2 | | Descriptor: | ACETATE ION, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | janocha, S, carius, y, bernhardt, r, lancaster, c.r.d. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
8B9O
 
 | |
8B9U
 
 | |
2KMU
 
 | |
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5LSS
 
 | |
5LT8
 
 | |
7XT6
 
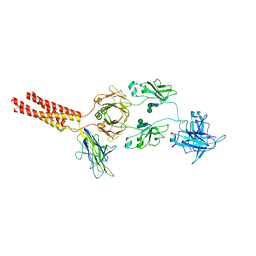 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
8EBU
 
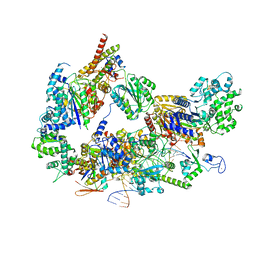 | | XPC release from Core7-XPA-DNA (Cy5) | | Descriptor: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBV
 
 | |
8EBS
 
 | |
8EBT
 
 | |
8EBW
 
 | |
8EBX
 
 | |
5LT7
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5LSX
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, ZINC ION, [(2~{R},5~{S})-1-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]-5-azaniumyl-6-oxidanyl-6-oxidanylidene-hexan-2-yl]-(phenylmethyl)azanium | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
5XUT
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
