4MW1
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1444) | | Descriptor: | 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4N5R
 
 | | Hen egg-white lysozyme phased using free-electron laser data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C | | Authors: | Barends, T.R.M, Foucar, L, Botha, S, Doak, R.B, Shoeman, R.L, Nass, K, Koglin, J.E, Williams, G.J, Boutet, S, Messerschmidt, M, Schlichting, I. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein crystal structure determination from X-ray free-electron laser data.
Nature, 505, 2014
|
|
3B5F
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating the Ade38Dap Mutation and a 2',5' Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
8VGQ
 
 | |
3BBK
 
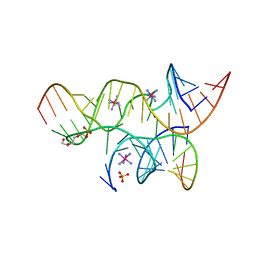 | | Miminally Junctioned Hairpin Ribozyme Incorporates A38C and 2'5'-phosphodiester Linkage within Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
8VGI
 
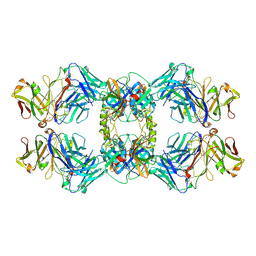 | |
8VEG
 
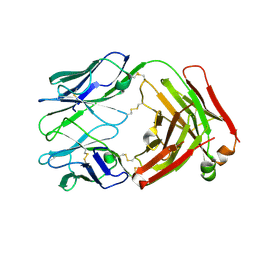 | |
8VGL
 
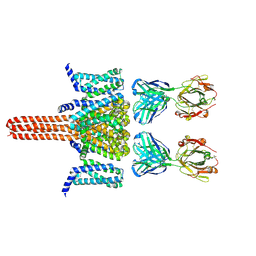 | | CryoEM structure of Nav1.7 in complex with wild type Fab 7A9 | | Descriptor: | Chimeric Nav1.7-NavAb, Fab 7A9 heavy chain, Fab 7A9 light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfi de constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
3BJI
 
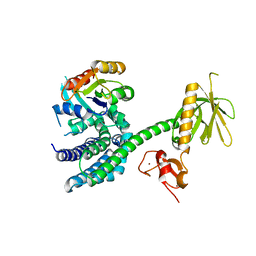 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
8VGE
 
 | |
8VGG
 
 | |
8VGP
 
 | |
8VGH
 
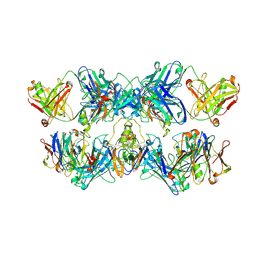 | |
8VGO
 
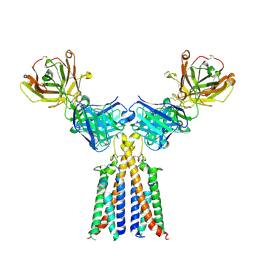 | | CryoEM structure of CD20 in complex with engineered conformationally rigid Rituximab.4DS Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab.4DS Fab heavy chain, Rituximab.4DS Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
8VGJ
 
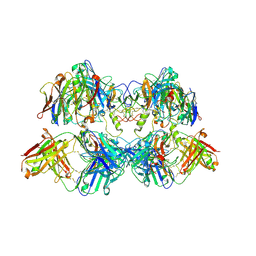 | |
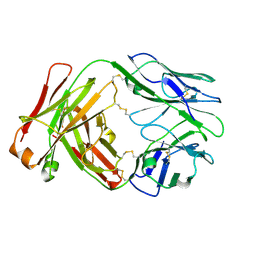
8VGK
 
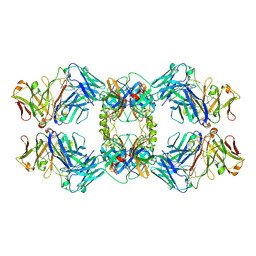 | |
8VGN
 
 | | CryoEM structure of CD20 in complex with wild type Rituximab Fab | | Descriptor: | B-lymphocyte antigen CD20, Rituximab Fab heavy chain, Rituximab Fab light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Disulfide constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
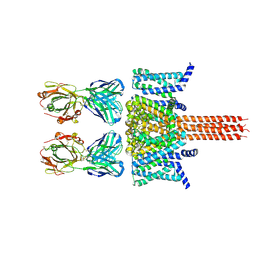
8VGF
 
 | |
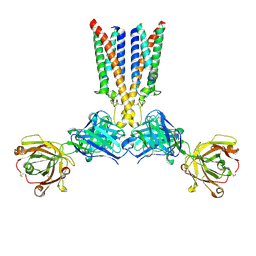
8VGM
 
 | | CryoEM structure of Nav1.7 in complex with engineered conformationally rigid Fab 7A9.4DS | | Descriptor: | Chimeric Nav1.7-NavAb, Fab 7A9.4DS heavy chain, Fab 7A9.4DS light chain | | Authors: | Kung, J.E, Jao, C.C, Arthur, C.P, Sudhamsu, J. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Disulfi de constrained Fabs overcome target size limitation for high-resolution single-particle cryo-EM.
Biorxiv, 2024
|
|
3BBI
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporating A38(2AP) and A-1 2'-O-Me Modifications near Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBM
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38C and 2'O-Me Modification at Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
2Y6S
 
 | | Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope | | Descriptor: | ENVELOPE GLYCOPROTEIN, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Olal, D.O, Kuehne, A, Lee, J.E, Bale, S, Dye, J.M, Saphire, E.O. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Ebola Virus-Protective Antibody in Complex with its Mucin-Domain Linear Epitope.
J.Virol., 86, 2012
|
|
4NKE
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Tsoumpra, M.K, Barnett, B.L, Muniz, J.R.C, Walter, R.L, Ebetino, F.H, von Delft, F, Russell, R.G.G, Oppermann, U, Dunford, J.E. | | Deposit date: | 2013-11-12 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
To be Published
|
|
2YCQ
 
 | | Crystal structure of checkpoint kinase 2 in complex with inhibitor PV1115 | | Descriptor: | N-{4-[(1E)-N-1H-IMIDAZOL-2-YLETHANEHYDRAZONOYL]PHENYL}-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
8VFM
 
 | |
