4O3S
 
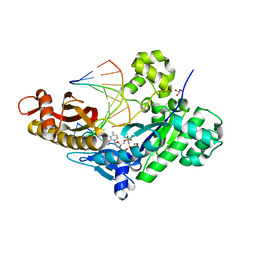 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-dc pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
3CIC
 
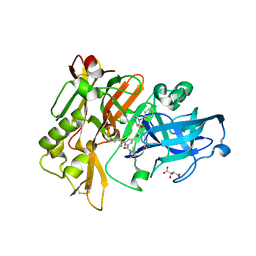 | | Structure of BACE Bound to SCH709583 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4OK5
 
 | |
5ULA
 
 | |
4P0O
 
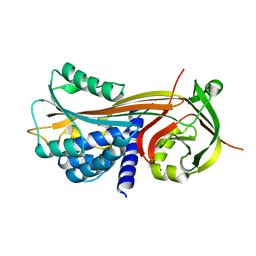 | | Cleaved Serpin 42Da | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
4P0F
 
 | | Cleaved Serpin 42Da (C 2 2 21) | | Descriptor: | Serine protease inhibitor 4, isoform B | | Authors: | Ellisdon, A.M, Whisstock, J.C. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster.
Bmc Struct.Biol., 14, 2014
|
|
4OKS
 
 | |
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MT4
 
 | |
4O3P
 
 | | Crystal structure of human polymerase eta inserting dctp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
8UVK
 
 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
4NRO
 
 | |
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4O3R
 
 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-da pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4NRP
 
 | |
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
4OJQ
 
 | |
4Q9J
 
 | | P-glycoprotein cocrystallised with QZ-Val | | Descriptor: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9H
 
 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QEC
 
 | | ElxO with NADP Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
