4HBX
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 3-methyl-6-(pyrrolidin-1-ylsulfonyl)-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
4HBV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
8B0V
 
 | | Crystal structure of C-terminal domain of Pseudomonas aeruginosa LexA G91D mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Vascon, F, De Felice, S, Chinellato, M, Maso, L, Cendron, L. | | Deposit date: | 2022-09-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigations on the SOS response in Pseudomonas aeruginosa
To Be Published
|
|
1GZ7
 
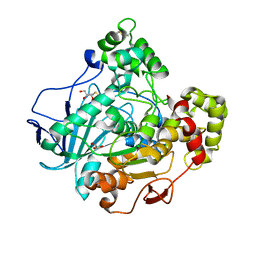 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
2IE2
 
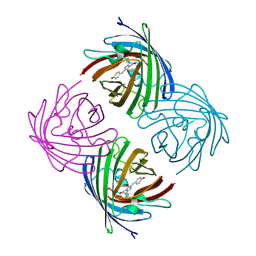 | |
3RXW
 
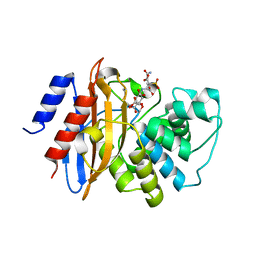 | | KPC-2 carbapenemase in complex with PSR3-226 | | Descriptor: | (2S,3R)-4-(2-amino-2-oxoethoxy)-3-(dihydroxy-lambda~4~-sulfanyl)-3-methyl-4-oxo-2-{[(1E)-3-oxoprop-1-en-1-yl]amino}butanoic acid, CITRIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2011-05-10 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
3RXX
 
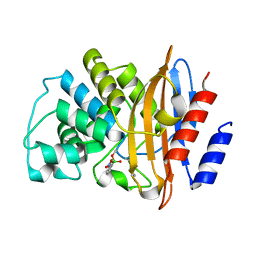 | | KPC-2 carbapenemase in complex with 3-NPBA | | Descriptor: | 3-NITROPHENYLBORONIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2011-05-10 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
2K06
 
 | |
2BSP
 
 | | BACILLUS SUBTILIS PECTATE LYASE R279K MUTANT | | Descriptor: | CALCIUM ION, PROTEIN (PECTATE LYASE) | | Authors: | Pickersgill, R. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Conserved Arginine Proximal to the Essential Calcium of Bacillus Subtilis Pectate Lyase Stabilizes the Transition State
To be Published
|
|
2JVV
 
 | |
2JUL
 
 | | NMR Structure of DREAM | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Ames, J. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization.
Biochemistry, 47, 2008
|
|
2G3O
 
 | | The 2.1A crystal structure of copGFP | | Descriptor: | green fluorescent protein 2 | | Authors: | Wilmann, P.G. | | Deposit date: | 2006-02-20 | | Release date: | 2006-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A crystal structure of copGFP, a representative member of the copepod clade within the green fluorescent protein superfamily
J.Mol.Biol., 359, 2006
|
|
2LMF
 
 | | Solution structure of human LL-23 bound to membrane-mimetic micelles | | Descriptor: | Antibacterial protein LL-37 | | Authors: | Wang, G. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Dynamics, and Antimicrobial and Immune Modulatory Activities of Human LL-23 and Its Single-Residue Variants Mutated on the Basis of Homologous Primate Cathelicidins.
Biochemistry, 51, 2012
|
|
3HFM
 
 | |
4Q7G
 
 | | 1.7 Angstrom Crystal Structure of leukotoxin LukD from Staphylococcus aureus. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leucotoxin LukDv | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Shatsman, S, Kwon, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1LY7
 
 | | The solution structure of the the c-terminal domain of frataxin, the protein responsible for friedreich ataxia | | Descriptor: | frataxin | | Authors: | Musco, G, Stier, G, Kolmerer, B, Adinolfi, S, Martin, S, Frenkiel, T, Gibson, T, Pastore, A. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards a structural understanding of Friedreich's
ataxia: the solution structure of frataxin
Structure Fold.Des., 8, 2000
|
|
4RWR
 
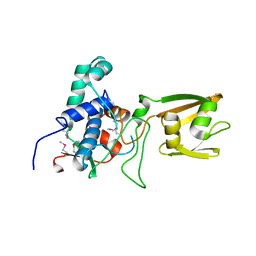 | | 2.1 Angstrom Crystal Structure of Stage II Sporulation Protein D from Bacillus anthracis | | Descriptor: | Stage II sporulation protein D | | Authors: | Minasov, G, Wawrzak, Z, Nocadello, S, Shuvalova, L, Dubrovska, I, Flores, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-05 | | Release date: | 2014-12-17 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the SpoIID Lytic Transglycosylases Essential for Bacterial Sporulation.
J.Biol.Chem., 291, 2016
|
|
1EP3
 
 | | CRYSTAL STRUCTURE OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE B. DATA COLLECTED UNDER CRYOGENIC CONDITIONS. | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE B (PYRD SUBUNIT), DIHYDROOROTATE DEHYDROGENASE B (PYRK SUBUNIT), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rowland, P, Norager, S, Jensen, K.F, Larsen, S. | | Deposit date: | 2000-03-27 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydroorotate dehydrogenase B: electron transfer between two flavin groups bridged by an iron-sulphur cluster.
Structure Fold.Des., 8, 2000
|
|
1ESK
 
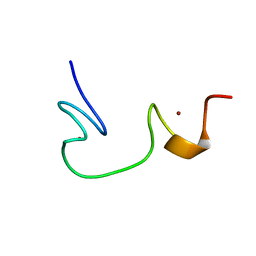 | | SOLUTION STRUCTURE OF NCP7 FROM HIV-1 | | Descriptor: | GAG POLYPROTEIN, ZINC ION | | Authors: | Morellet, N, Demene, H, Teilleux, V, Huynh-Dinh, T, de Rocquigny, H, Fournie-Zaluski, M.-C, Roques, B.P. | | Deposit date: | 2000-04-10 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of (12-53)NCp7 of HIV-1
To be Published
|
|
3ROH
 
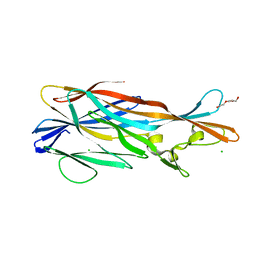 | | Crystal Structure of Leukotoxin (LukE) from Staphylococcus aureus subsp. aureus COL. | | Descriptor: | CHLORIDE ION, Leucotoxin LukEv, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2VDG
 
 | | Barley Aldose Reductase 1 complex with butanol | | Descriptor: | 1-BUTANOL, ALDOSE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2007-10-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
