4HBC
 
 | |
3HWO
 
 | |
5JPX
 
 | |
6GMA
 
 | |
6NMC
 
 | | CryoEM structure of the LbCas12a-crRNA-2xAcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NMD
 
 | | cryo-EM Structure of the LbCas12a-crRNA-AcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
5XVR
 
 | |
6UH7
 
 | | EV-A71 strain 11316 complexed with MADAL compound 30 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
6UH6
 
 | | EV-A71 strain 11316 complexed with MADAL compound 22 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1GU0
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Krell, T, Robinson, D, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
5WXI
 
 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXK
 
 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
1GTZ
 
 | | Structure of STREPTOMYCES COELICOLOR TYPE II DEHYDROQUINASE R23A MUTANT IN COMPLEX WITH DEHYDROSHIKIMATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, 3-DEHYDROSHIKIMATE | | Authors: | Roszak, A.W, Krell, T, Robinson, D.A, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
6VS9
 
 | |
6UH1
 
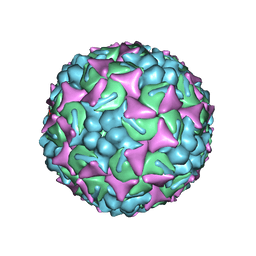 | | Structure of the EVA71 strain 11316 capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VS5
 
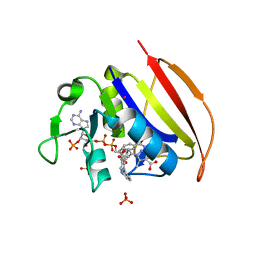 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-methyl-1-phenyl-1H-pyrazole-4-carboxylic acid (fragment 1) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Tyrakis, P, Blundell, T, Dias, M.V.B. | | Deposit date: | 2020-02-10 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VVB
 
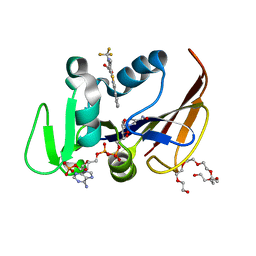 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 6-methyl-5-(4-phenylthiazol-2-yl)-2- (trifluoromethyl)nicotinic acid (fragment 10) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 6-methyl-5-(4-phenyl-1,3-thiazol-2-yl)-2-(trifluoromethyl)pyridine-3-carboxylic acid, Dihydrofolate reductase, ... | | Authors: | Tyrakis, P, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VSE
 
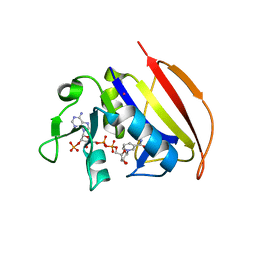 | |
6VV9
 
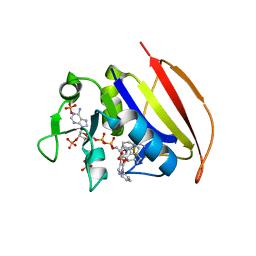 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB300 | | Descriptor: | 5-[4-(1H-indol-3-yl)butoxy]-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VSD
 
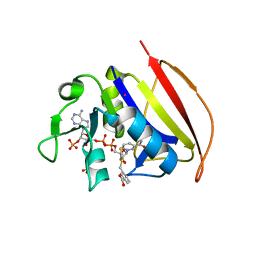 | |
6VSG
 
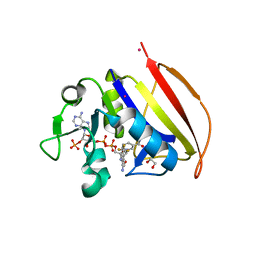 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(trifluoromethyl)benzene-1,2-diamine(fragment 17) | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VSF
 
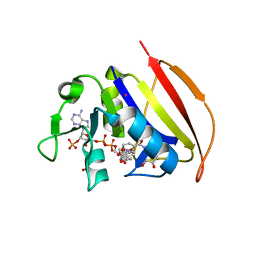 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(3,4-dihydro-2H-benzo[b][1,4]dioxepin-7-yl)-4-oxobutanoic acid(fragment 16) | | Descriptor: | 4-(3,4-dihydro-2H-1,5-benzodioxepin-7-yl)-4-oxobutanoic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VS8
 
 | |
