5Y8Z
 
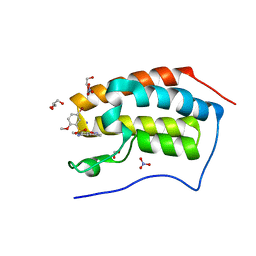 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
1ZXC
 
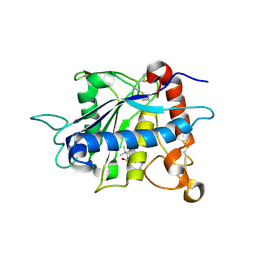 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | Descriptor: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1ZYV
 
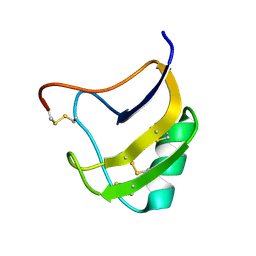 | | Crystal Structure Of Mutant K8DP9SR58KV59G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
1ZUT
 
 | | Crystal Structure Of Mutant K8DP9SR58K Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZYW
 
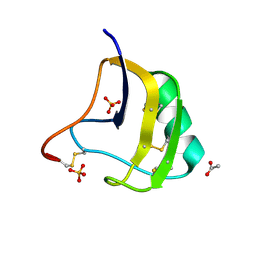 | | Crystal Structure Of Mutant K8DP9SR58KP60G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1MSV
 
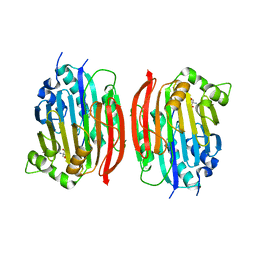 | | The S68A S-adenosylmethionine decarboxylase proenzyme processing mutant. | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Tolbert, W.D, Zhang, Y, Bennett, E.M, Cottet, S.E, Ekstrom, J.L, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of Human S-Adenosylmethionine
Decarboxylase Proenzyme Processing as Revealed by the
Structure of the S68A Mutant.
Biochemistry, 42, 2003
|
|
7AW0
 
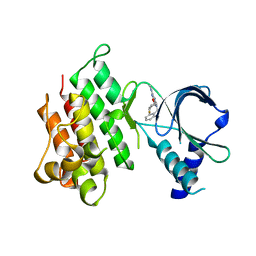 | | MerTK kinase domain in complex with purine inhibitor | | Descriptor: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
5KPV
 
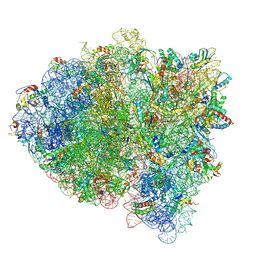 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPW
 
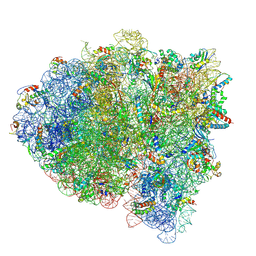 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6DNH
 
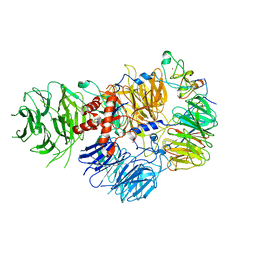 | | Cryo-EM structure of human CPSF-160-WDR33-CPSF-30-PAS RNA complex at 3.4 A resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*C)-3'), ... | | Authors: | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5KPS
 
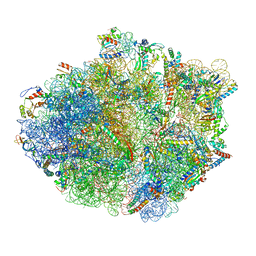 | | Structure of RelA bound to ribosome in absence of A/R tRNA (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
1ZCN
 
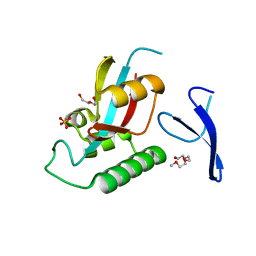 | | human Pin1 Ng mutant | | Descriptor: | PENTAETHYLENE GLYCOL, PHOSPHATE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Nguyen, H, Dendel, G, Bowman, M.E, Gruebele, M, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-04-12 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1L3K
 
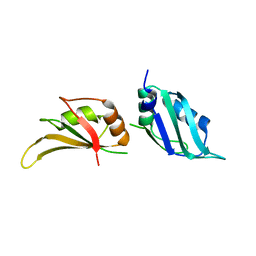 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
5LNU
 
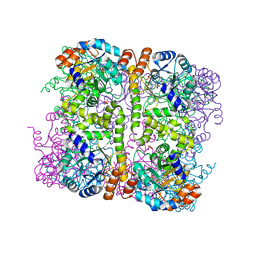 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNS
 
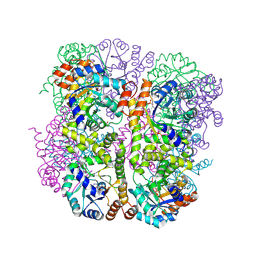 | | Crystal structure of Arabidopsis thaliana Pdx1-R5P complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, RIBULOSE-5-PHOSPHATE | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
3JA8
 
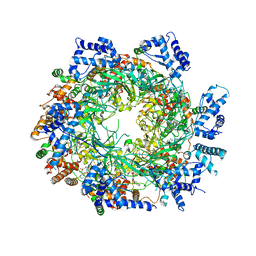 | | Cryo-EM structure of the MCM2-7 double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | Authors: | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
4U86
 
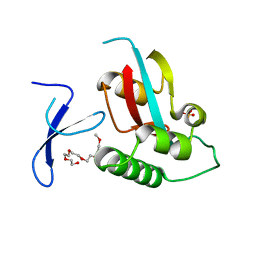 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
4U85
 
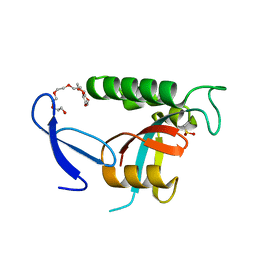 | | Human Pin1 with cysteine sulfinic acid 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
1Z68
 
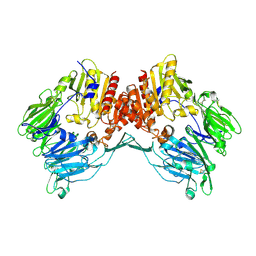 | | Crystal Structure Of Human Fibroblast Activation Protein alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aertgeerts, K, Levin, I, Shi, L, Prasad, G.S, Zhang, Y, Kraus, M.L, Salakian, S, Snell, G.P, Sridhar, V, Wijnands, R, Tennant, M.G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha.
J.Biol.Chem., 280, 2005
|
|
5LNW
 
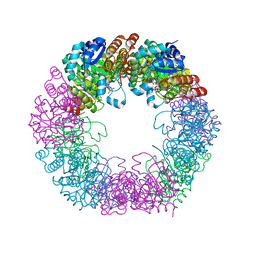 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
1UNX
 
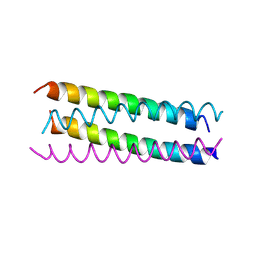 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UO1
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UO2
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
