1MUZ
 
 | |
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
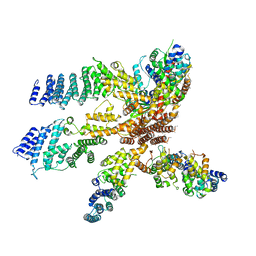 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
1MV0
 
 | |
2JZT
 
 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
2YGW
 
 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
