3DT5
 
 | | C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Uncharacterized protein AF_0924 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-14 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray crystal structure of C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus.
To be Published
|
|
3HFU
 
 | | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, HTH-type transcriptional regulator cynR | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Dong, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide
TO BE PUBLISHED
|
|
3HHQ
 
 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
4MND
 
 | | Crystal structure of Archaeoglobus fulgidus IPCT-DIPPS bifunctional membrane protein | | Descriptor: | CTP L-myo-inositol-1-phosphate cytidylyltransferase/CDP-L-myo-inositol myo-inositolphosphotransferase, EICOSANE, MAGNESIUM ION | | Authors: | Nogly, P, Gushchin, I, Remeeva, A, Esteves, A.M, Ishchenko, A, Ma, P, Grudinin, S, Borges, N, Round, E, Moraes, I, Borshchevskiy, V, Santos, H, Gordeliy, V, Archer, M. | | Deposit date: | 2013-09-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism.
Nat Commun, 5, 2014
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3C9H
 
 | | Crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens | | Descriptor: | ABC transporter, substrate binding protein, CITRIC ACID, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the substrate binding protein of the ABC transporter from Agrobacterium tumefaciens.
To be Published
|
|
3CBT
 
 | | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor | | Descriptor: | MAGNESIUM ION, Phosphatase SC4828, SODIUM ION | | Authors: | Singer, A.U, Xu, X, Chang, C, Zheng, H, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SC4828, a unique phosphatase from Streptomyces coelicolor.
To be Published
|
|
3CDD
 
 | | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis | | Descriptor: | Prophage MuSo2, 43 kDa tail protein | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prophage MuSo2, 43 kDa tail protein from Shewanella oneidensis.
To be Published
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3C5O
 
 | | Crystal structure of the conserved protein of unknown function RPA1785 from Rhodopseudomonas palustris | | Descriptor: | GLYCEROL, UPF0311 protein RPA1785 | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Conserved Protein of Unknown Function RPA1785 from Rhodopseudomonas palustris.
To be Published
|
|
2IBD
 
 | | Crystal structure of Probable transcriptional regulatory protein RHA5900 | | Descriptor: | MAGNESIUM ION, Possible transcriptional regulator | | Authors: | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Probable transcriptional regulatory protein RHA5900
To be Published
|
|
4XV0
 
 | | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei | | Descriptor: | Beta-xylanase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9697 Å) | | Cite: | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei
To Be Published
|
|
4XX6
 
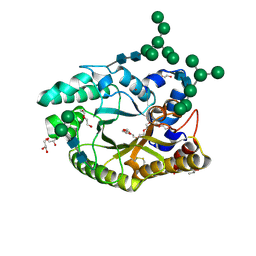 | | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Stogios, P.J, Nocek, B, Xu, X, Cui, H, Lowden, M, Savchenko, A. | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum.
To Be Published
|
|
3ICF
 
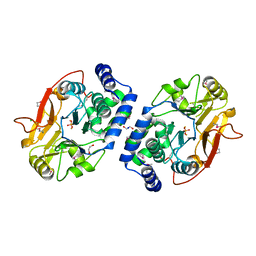 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
3CQY
 
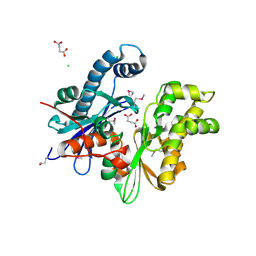 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
3CUO
 
 | | Crystal structure of the predicted DNA-binding transcriptional regulator from E. coli | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygaV | | Authors: | Zhang, R, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the predicted DNA-binding transcriptional regulator from E. coli.
To be Published
|
|
3FYN
 
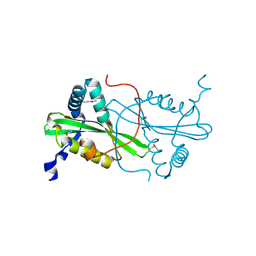 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3L9A
 
 | | Structure of the C-terminal domain from a Streptococcus mutans hypothetical | | Descriptor: | GLYCEROL, SODIUM ION, uncharacterized protein | | Authors: | Singer, A.U, Cuff, M.E, Xu, X, Cui, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-04 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the C-terminal domain from a Streptococcus mutans hypothetical
To be Published
|
|
4XUV
 
 | | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris | | Descriptor: | GLYCEROL, Glycoside hydrolase family 105 protein | | Authors: | Stogios, P.J, Xu, X, Cui, H, Yim, V, Savchenko, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0496 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 105 (GH105) enzyme from Thielavia terrestris
To Be Published
|
|
5BQ9
 
 | | Crystal structure of uncharacterized protein lpg1496 Legionella pneumophila subsp. pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Morar, M, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2785 Å) | | Cite: | Crystal structure of the Legionella pneumophila lem10 effector reveals a new member of the HD protein superfamily.
Proteins, 83, 2015
|
|
1XSV
 
 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | Hypothetical UPF0122 protein SAV1236 | | Authors: | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
3IB3
 
 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
3III
 
 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
