5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
6B0Y
 
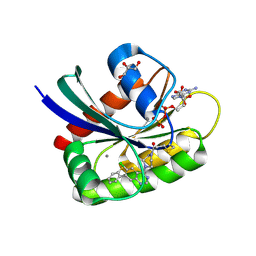 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8K58
 
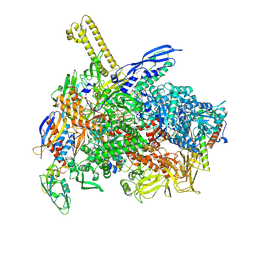 | | The cryo-EM map of close TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TIC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
6D2V
 
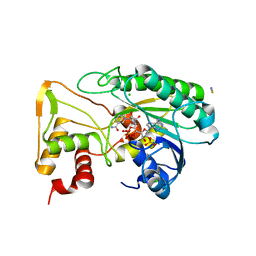 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6D34
 
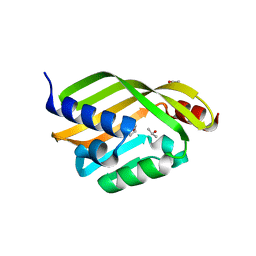 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
2FTP
 
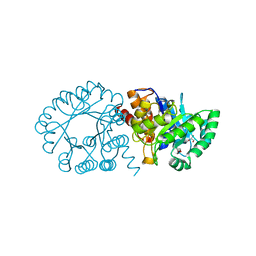 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
5TTH
 
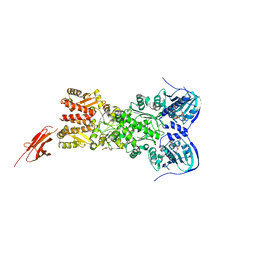 | | Heterodimeric SpyCatcher/SpyTag-fused zebrafish TRAP1 in ATP/ADP-hybrid state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, C-terminal SpyCatcher fusion of wildtype zebrafish TNF receptor-associated protein 1, ... | | Authors: | Elnatan, D, Betegon, M, Liu, Y, Agard, D.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1.
Elife, 6, 2017
|
|
3LNW
 
 | |
3EBJ
 
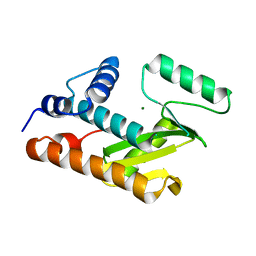 | | Crystal structure of an avian influenza virus protein | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein | | Authors: | Yuan, P, Bartlam, M, Lou, Z, Chen, S, Rao, Z, Liu, Y. | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site
Nature, 458, 2009
|
|
2H40
 
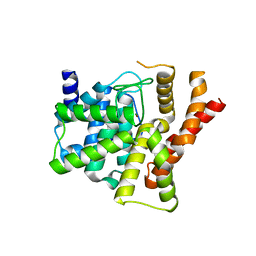 | | Crystal structure of the catalytic domain of unliganded PDE5 | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Liu, Y, Huai, Q, Cai, J, Zoraghi, R, Francis, S.H, Corbin, J.D, Robinson, H, Xin, Z, Lin, G, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
1ZKN
 
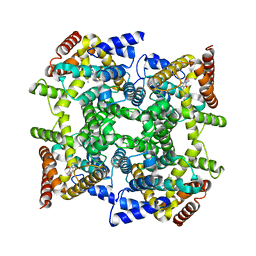 | | Structure of PDE4D2-IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Liu, Y, Francis, S.H, Corbin, J.D, Ke, H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Phosphodiesterases 4 and 5 in Complex with Inhibitor 3-Isobutyl-1-Methylxanthine Suggest a Conformation Determinant of Inhibitor Selectivity
J.Biol.Chem., 279, 2004
|
|
1YP1
 
 | | Crystal structure of a non-hemorrhagic fibrin(ogen)olytic metalloproteinase from venom of Agkistrodon acutus | | Descriptor: | FII, KNL, ZINC ION | | Authors: | Lou, Z, Hou, J, Chen, J, Liang, X, Qiu, P, Liu, Y, Li, M, Rao, Z. | | Deposit date: | 2005-01-28 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a non-hemorrhagic fibrin(ogen)olytic metalloproteinase complexed with a novel natural tri-peptide inhibitor from venom of Agkistrodon acutus
J.Struct.Biol., 152, 2005
|
|
3R7W
 
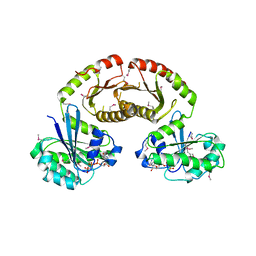 | | Crystal Structure of Gtr1p-Gtr2p complex | | Descriptor: | GTP-binding protein GTR1, GTP-binding protein GTR2, MAGNESIUM ION, ... | | Authors: | Gong, R, Li, L, Liu, Y, Wang, P, Yang, H, Wang, L, Cheng, J, Guan, K.L, Xu, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-08-24 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation
Genes Dev., 25, 2011
|
|
4QN7
 
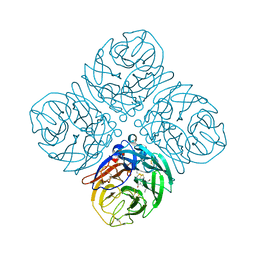 | | Crystal structure of neuramnidase N7 complexed with Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
5H3V
 
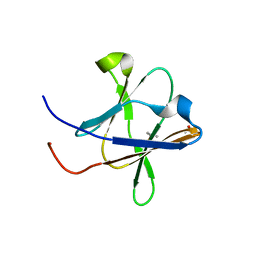 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3SAL
 
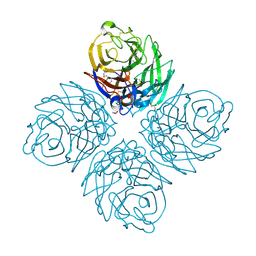 | | Crystal Structure of Influenza A Virus Neuraminidase N5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, M.Y, Qi, J.X, Liu, Y, Vavricka, C.J, Wu, Y, Li, Q, Gao, G.F. | | Deposit date: | 2011-06-03 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza a virus n5 neuraminidase has an extended 150-cavity
J.Virol., 85, 2011
|
|
4QN3
 
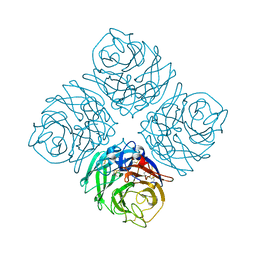 | | Crystal structure of Neuraminidase N7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
1Y4U
 
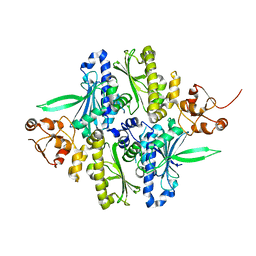 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
2APQ
 
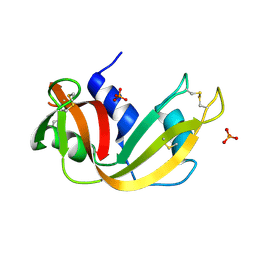 | | Crystal Structure of an Active Site Mutant of Bovine Pancreatic Ribonuclease A (H119A-RNase A) with a 10-Glutamine expansion in the C-terminal hinge-loop. | | Descriptor: | PHOSPHATE ION, Ribonuclease | | Authors: | Sambashivan, S, Liu, Y, Sawaya, M.R, Gingery, M, Eisenberg, D. | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure.
Nature, 437, 2005
|
|
1Y4E
 
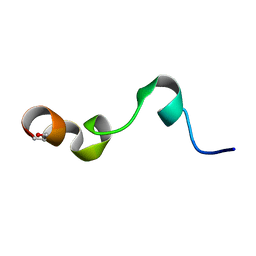 | | NMR structure of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Slepkov, E.R, Rainey, J.K, Li, X, Liu, Y, Lindhout, D.A, Sykes, B.D, Fliegel, L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger.
J.Biol.Chem., 280, 2005
|
|
1Y4S
 
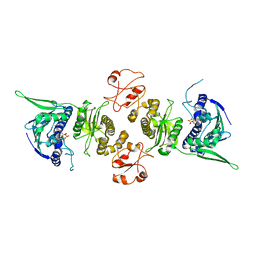 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
4QN5
 
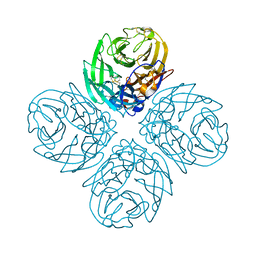 | | Neuraminidase N5 binds LSTa at the second SIA binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
