4YHS
 
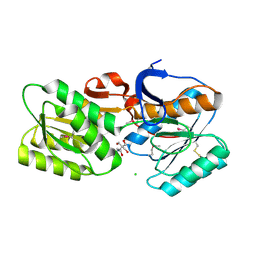 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BRADYRHIZOBIUM sp. BTAi1 (BBta_2440, TARGET EFI-511490) WITH BOUND BIS-TRIS
To be published
|
|
4YKY
 
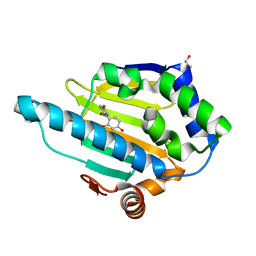 | | Heat Shock Protein 90 Bound to CS319 | | Descriptor: | (2,4-dihydroxyphenyl)(4-hydroxyphenyl)methanone, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS319
To Be Published
|
|
4Y64
 
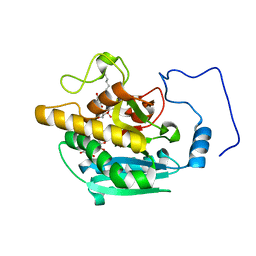 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-(D-alanylamino)-5'-deoxyuridine, Histo-blood group ABO system transferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
4Y72
 
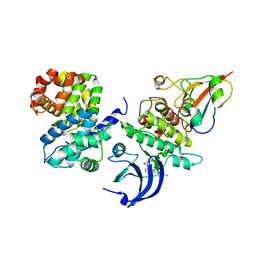 | | Human CDK1/CyclinB1/CKS2 With Inhibitor | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4Y9T
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GLUCOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, ABC transporter, solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YFT
 
 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YKZ
 
 | | Heat Shock Protein 90 Bound to CS320 | | Descriptor: | (2,4-dihydroxyphenyl)(3-hydroxyphenyl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS320
To Be Published
|
|
4YQM
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-27 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-N-[4-chloro-3-(dimethylsulfamoyl)phenyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YKT
 
 | | Heat Shock Protein 90 Bound to CS307 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-5-({[dihydroxy(pyridin-3-yl)-lambda~4~-sulfanyl]amino}methyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS307
To Be Published
|
|
4YQQ
 
 | |
4YWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE | | Descriptor: | ABC TRANSPORTER SOLUTE BINDING PROTEIN, beta-D-xylopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM ACTINOBACILLUS SUCCINOGENES 130Z (Asuc_0499, TARGET EFI-511068) WITH BOUND D-XYLOSE
To be published
|
|
4YIC
 
 | | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID | | Descriptor: | ACETATE ION, CALCIUM ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-01 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM BORDETELLA BRONCHISEPTICA RB50 (BB0280, TARGET EFI-500035) WITH BOUND PICOLINIC ACID
To be published
|
|
4YKW
 
 | | Heat Shock Protein 90 Bound to CS312 | | Descriptor: | 4-(2-chloro-4-nitrophenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS312
To Be Published
|
|
4YOE
 
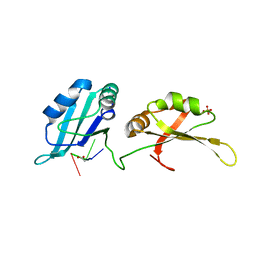 | | Structure of UP1 bound to RNA 5'-AGU-3' | | Descriptor: | ACETATE ION, Heterogeneous nuclear ribonucleoprotein A1, RNA AGU, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The First Crystal Structure of the UP1 Domain of hnRNP A1 Bound to RNA Reveals a New Look for an Old RNA Binding Protein.
J.Mol.Biol., 427, 2015
|
|
4YEW
 
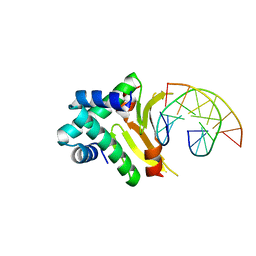 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YQY
 
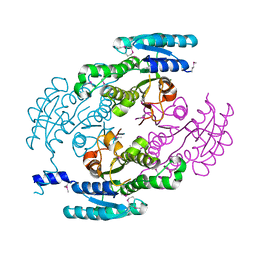 | | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028) (TARGET EFI-513936) in its APO form | | Descriptor: | MAGNESIUM ION, Putative Dehydrogenase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028, TARGET EFI-513936) in its APO form
To be published
|
|
4YS3
 
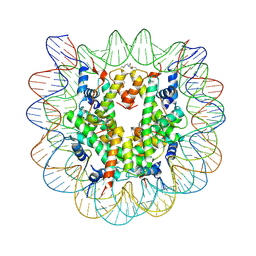 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
4YQV
 
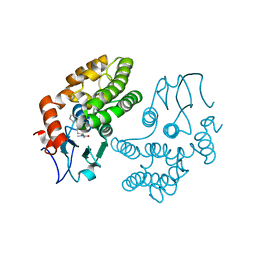 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C4-10 | | Descriptor: | 2-(ethylsulfanyl)-N-methyl-N-[(1-phenyl-1H-pyrazol-4-yl)methyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YS6
 
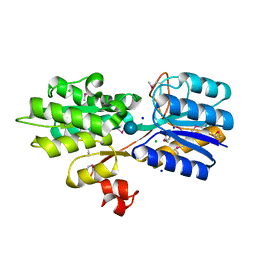 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE | | Descriptor: | CHLORIDE ION, Putative solute-binding component of ABC transporter, SODIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTANS (Cphy_1585, TARGET EFI-511156) WITH BOUND BETA-D-GLUCOSE
To be published
|
|
4YW5
 
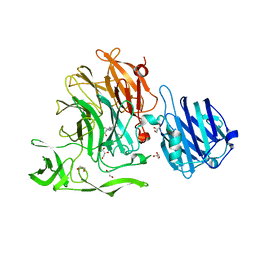 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4Y63
 
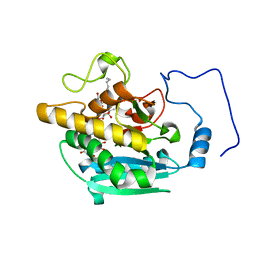 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2S)-2-(triaza-1,2-dien-2-ium-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
4YC3
 
 | | CDK1/CyclinB1/CKS2 Apo | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4YEY
 
 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4Y62
 
 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2R)-3-hydroxy-2-(4-phenyl-1H-1,2,3-triazol-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
