7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
5XYL
 
 | |
6K26
 
 | | Crystal structure of Vibrio cholerae methionine aminopeptidase | | Descriptor: | Methionine aminopeptidase, SODIUM ION | | Authors: | Pillalamarri, V, Addlagatta, A. | | Deposit date: | 2019-05-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Methionine aminopeptidases with short sequence inserts within the catalytic domain are differentially inhibited: Structural and biochemical studies of three proteins from Vibrio spp.
Eur.J.Med.Chem., 209, 2020
|
|
7CEF
 
 | | Crystal structure of PET-degrading cutinase Cut190 /S226P/R228S/ mutant with the C-terminal three residues deletion | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, ZINC ION | | Authors: | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
8QPH
 
 | | Crystal structure of Lymantria dispar CPV14 polyhedra 14 crystals | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
7CBS
 
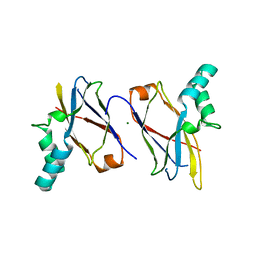 | | Crystal structure of SpaB basal pilin from Lactobacillus rhamnosus GG | | Descriptor: | CHLORIDE ION, LPXTG cell wall anchor domain-containing protein, MAGNESIUM ION | | Authors: | Megta, A.K, Pratap, S, Kant, A, Krishnan, V. | | Deposit date: | 2020-06-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the atypically adhesive SpaB basal pilus subunit: Mechanistic insights about its incorporation in lactobacillar SpaCBA pili.
Curr Res Struct Biol, 2, 2020
|
|
6JT4
 
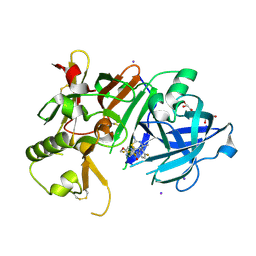 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
7CEH
 
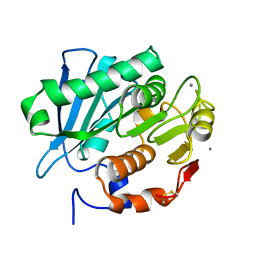 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant with the C-terminal three residues deletion in ligand ejecting form | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Senga, A, Numoto, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-06-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Multiple structural states of Ca2+-regulated PET hydrolase, Cut190, and its correlation with activity and stability.
J.Biochem., 169, 2021
|
|
8QQC
 
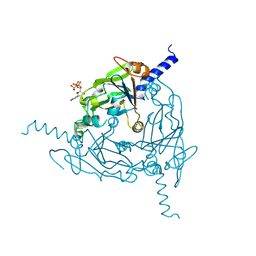 | | Crystal structure of Lymantria dispar CPV14 polyhedra single crystal | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Trincao, J, Warren, A, Crawshaw, A, Sutton, G, Stuart, D, Evans, G. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | VMXm - sub-micron microfocus beamline for macromolecular crystallography at Diamond Light Source
To Be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
3CNG
 
 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3CP8
 
 | | Crystal structure of GidA from Chlorobium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Meyer, S, Scrima, A, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
6GW9
 
 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
5Z47
 
 | | Crystal structure of pyrrolidone carboxylate peptidase I with disordered loop A from Deinococcus radiodurans R1 | | Descriptor: | DIMETHYL SULFOXIDE, Pyrrolidone-carboxylate peptidase | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of pyrrolidone-carboxylate peptidase I from Deinococcus radiodurans reveal the mechanism of L-pyroglutamate recognition.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3D1P
 
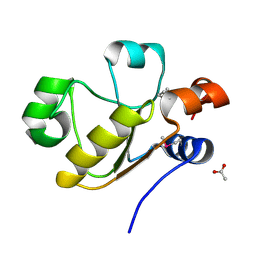 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
8UPT
 
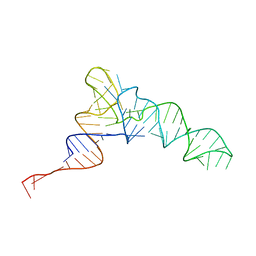 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
6KKV
 
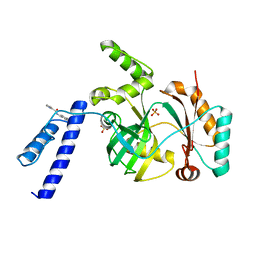 | |
3CG9
 
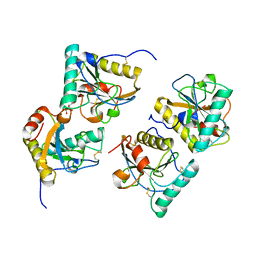 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
7QZP
 
 | | Identification and characterization of an RRM-containing, ELAV-like, RNA binding protein in Acinetobacter Baumannii | | Descriptor: | Hypothetical RNA binding protein from Acinetobacter baumannii | | Authors: | Ciani, C, Perez-Rafols, A, Bonomo, I, Micaelli, M, Esposito, A, Zucal, C, Belli, R, D'Agostino, V.G, Bianconi, I, Calderone, V, Cerofolini, L, Fragai, M, Provenzani, A. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii .
Biomolecules, 12, 2022
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
