7MQI
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MQG
 
 | | Bartonella henselae NrnC complexed with pAAAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPM
 
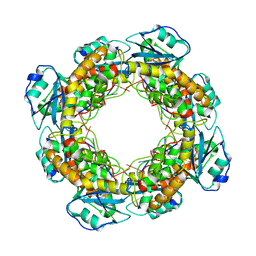 | | Bartonella henselae NrnC bound to pAA | | Descriptor: | 5'-phosphorylated AA, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPN
 
 | | Bartonella henselae NrnC bound to pGC | | Descriptor: | 5'-phosphorylated GC, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPQ
 
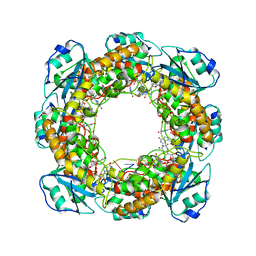 | |
7MQH
 
 | | Bartonella henselae NrnC complexed with pAAAGG in the presence of Ca2+. D4 Symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*AP*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MFG
 
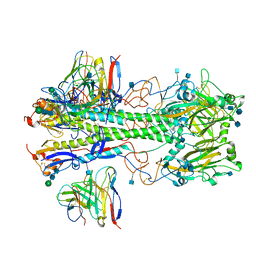 | | Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 310-030-1D06 Heavy, 310-030-1D06 Light, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-04-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
7MGK
 
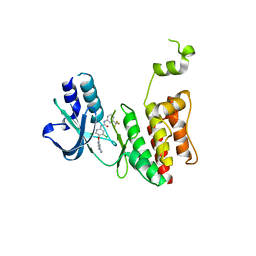 | |
3U0J
 
 | |
7MGJ
 
 | |
2LH0
 
 | |
4EQZ
 
 | | Crystal structure of human DOT1L in complex with inhibitor FED2 | | Descriptor: | 5'-deoxy-5'-[(3-{[(4-methylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]adenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
7MT4
 
 | | Crystal structure of tryptophan Synthase in complex with F9, NH4+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMMONIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT5
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, CESIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4EG7
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1331 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, METHIONINE, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
7MT6
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, benzimidazole, pH7.8 - alpha aminoacrylate form - E(A-A)(BZI) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4EI4
 
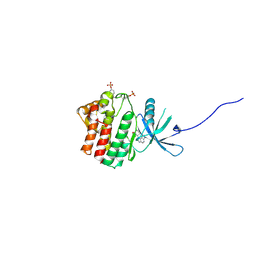 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4EM4
 
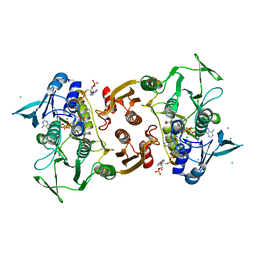 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor Pethyl-VS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4EG5
 
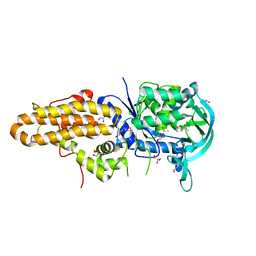 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor Chem 1312 | | Descriptor: | 2-({3-[(3,5-dichlorobenzyl)amino]propyl}amino)quinolin-4(1H)-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Shibata, S, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection.
Structure, 20, 2012
|
|
4ER0
 
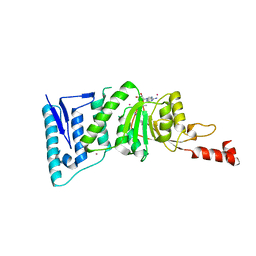 | | Crystal Structure of human DOT1L in complex with inhibitor FED1 | | Descriptor: | 5'-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5'-deoxyadenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER7
 
 | | Crystal Structure of human DOT1L in complex with inhibitor SGC0947 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
3V4J
 
 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | 4-[methyl(nitroso)amino]benzene-1,2-diol, DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
4EM3
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor MeVS-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wallace, B.D, Edwards, J.S, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-11 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
2LCG
 
 | | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans, Northeast Structural Genomics Consortium Target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Yang, Y, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans.
Northeast Structural Genomics Consortium Target CrR115.
To be Published
|
|
3V6E
 
 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
