8U9L
 
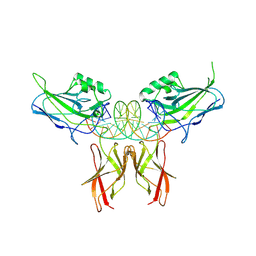 | | Crystal Structure of RelA-cRel chimera complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*AP*TP*TP*C)-3'), Transcription factor p65,Proto-oncogene c-Rel chimera | | Authors: | Chang, A, Wu, Y, Li, S.X, Smale, S, Chen, L. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Stepwise neofunctionalization of the NF-kappa B family member Rel during vertebrate evolution.
Nat.Immunol., 26, 2025
|
|
2DUY
 
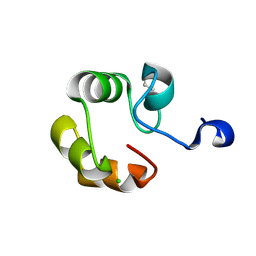 | | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Competence protein ComEA-related protein | | Authors: | Niwa, H, Shimada, A, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8
To be Published
|
|
2PH3
 
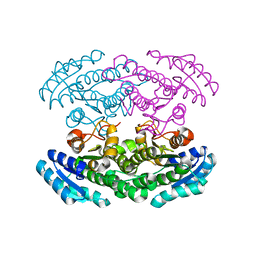 | | Crystal structure of 3-oxoacyl-[acyl carrier protein] reductase TTHA0415 from Thermus thermophilus | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase | | Authors: | Swindell II, J.T, Chen, L, Zhu, J, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of 3-oxoacyl-[acyl carrier protein] reductase TTHA0415 from Thermus thermophilus
To be Published
|
|
2P5U
 
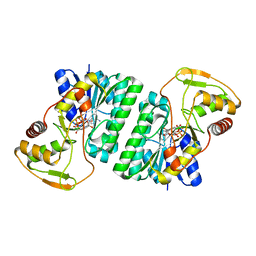 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P9M
 
 | | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Hypothetical protein MJ0922 | | Authors: | Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell II, J.T, Chen, L, Fu, Z.-Q, Charz, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
8VAD
 
 | | Crystal structure of MCoHNE-I, a potent in-vivo neutrophil elastase inhibitor | | Descriptor: | SULFATE ION, Trypsin inhibitor 2 | | Authors: | Xu, J, Rajasekaran, G, Li, S, Chen, L, Camarero, J.A. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of MCoHNE-I, a potent in-vivo neutrophil elastase inhibitor
To Be Published
|
|
8VK5
 
 | | Human Sputum Leucocyte Elastase (uncomplexed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Neutrophil elastase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xu, J, Rajasekaran, G, Li, S, Chen, L, Camarero, J.A. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Human Sputum Leucocyte Elastase (uncomplexed)
To Be Published
|
|
8VY8
 
 | | Recombinant alpha bungarotoxin complexed with HAP peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alpha-bungarotoxin isoform V31, HAP peptide | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Scalable production of recombinant three-finger proteins: from inclusion bodies to high quality molecular probes.
Microb Cell Fact, 23, 2024
|
|
2FAT
 
 | | An anti-urokinase plasminogen activator receptor (UPAR) antibody: Crystal structure and binding epitope | | Descriptor: | FAB ATN-615, heavy chain, light chain | | Authors: | Li, Y, Parry, G, Shi, X, Chen, L, Callahan, J.A, Mazar, A.P, Huang, M. | | Deposit date: | 2005-12-07 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An anti-urokinase plasminogen activator receptor (uPAR) antibody: crystal structure and binding epitope
J.Mol.Biol., 365, 2007
|
|
2O93
 
 | |
2P5Y
 
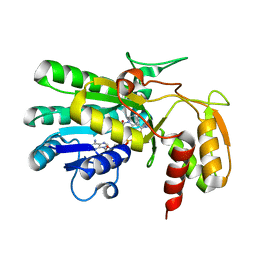 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P3E
 
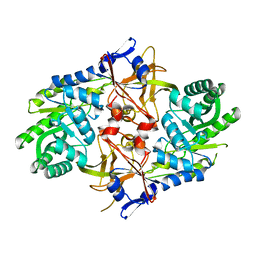 | | Crystal structure of AQ1208 from Aquifex aeolicus | | Descriptor: | Diaminopimelate decarboxylase | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-08 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be Published
To be Published
|
|
2P9J
 
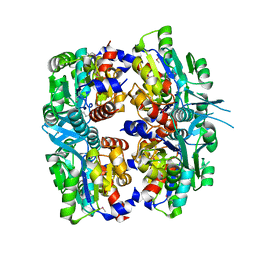 | | Crystal structure of AQ2171 from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ2171 | | Authors: | Yang, H, Chen, L, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AQ2171 from Aquifex aeolicus
To be Published
|
|
2PK8
 
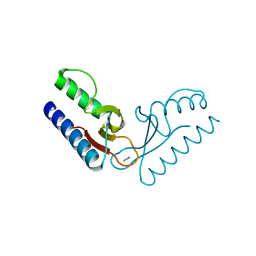 | | Crystal structure of an uncharacterized protein PF0899 from Pyrococcus furiosus | | Descriptor: | GOLD (I) CYANIDE ION, Uncharacterized protein PF0899 | | Authors: | Liu, Z.J, Tempel, W, Chen, L, Shah, A, Lee, D, Clancy-Kelley, L.L, Dillard, B.D, Rose, J.P, Sugar, F.J, Jenny Jr, F.E, Lee, H.S, Izumi, M, Shah, C, Poole III, F.L, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2QVO
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
2QC1
 
 | | Crystal structure of the extracellular domain of the nicotinic acetylcholine receptor 1 subunit bound to alpha-bungarotoxin at 1.9 A resolution | | Descriptor: | Acetylcholine receptor subunit alpha, Alpha-bungarotoxin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Dellisanti, C.D, Yao, Y, Stroud, J.C, Wang, Z, Chen, L. | | Deposit date: | 2007-06-18 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution.
Nat.Neurosci., 10, 2007
|
|
2PW6
 
 | | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12 | | Descriptor: | Uncharacterized protein ygiD, ZINC ION | | Authors: | Newton, M.G, Takagi, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokayama, S, Li, Y, Chen, L, Zhu, J, Ruble, J, Liu, Z.J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12.
To be Published
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
2E7V
 
 | | Crystal structure of SEA domain of transmembrane protease from Mus musculus | | Descriptor: | Transmembrane protease | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Shirozu, M, Yokoyama, S, Chen, L, Liu, Z.J, Wang, B.C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SEA domain of transmembrane protease from Mus musculus
To be Published
|
|
2E1D
 
 | | Crystal structure of mouse transaldolase | | Descriptor: | SULFITE ION, Transaldolase | | Authors: | Kishishita, S, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
2EIS
 
 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2P17
 
 | | Crystal structure of GK1651 from Geobacillus kaustophilus | | Descriptor: | FE (III) ION, Pirin-like protein | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of GK1651 from Geobacillus kaustophilus
To be Published
|
|
2DYY
 
 | | Crystal structure of putative translation initiation inhibitor PH0854 from Pyrococcus horikoshii | | Descriptor: | UPF0076 protein PH0854 | | Authors: | Ihsanawati, Kishishita, S, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-19 | | Release date: | 2007-03-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative translation initiation inhibitor PH0854 from Pyrococcus horikoshii
To be Published
|
|
2P2O
 
 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
2E0T
 
 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | Descriptor: | Dual specificity phosphatase 26 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
